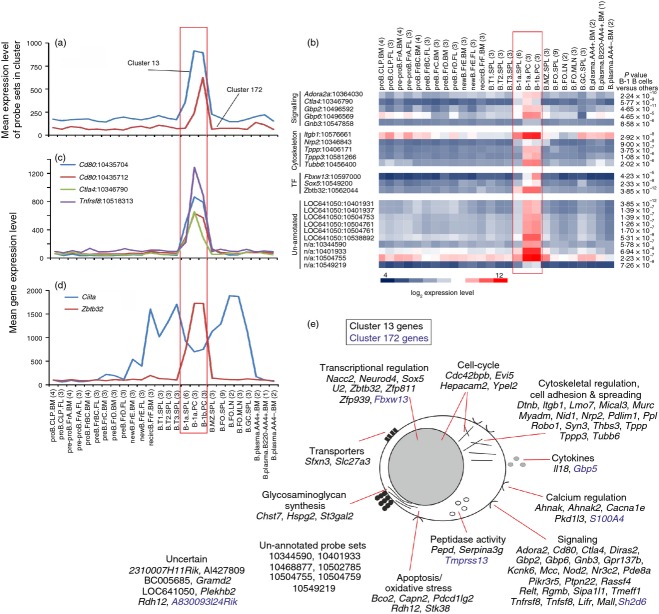
Figure 4.
Analysis of the genes within clusters 13 and 172 which were expressed specifically by B1 B cells. (a) The mean expression profile of all the probe set intensities within clusters 13 (blue) and 172 (red) over the 84 samples. (b) Heat map showing the mean expression levels of probe sets of interest in clusters 13 and 172. Each column represents the mean (log2) probe set intensity for all samples from each source. Significant differences between groups were sought by analysis of variance. P-values for those genes that were expressed significantly (P < 0·05) by B1 B cells at levels > 2·0 fold when compared with the other cell populations. (c) The mean expression profile of probe sets representing Ctla4 (green), Cd80 (red and blue) and Tnfrsf8 (purple) across the 84 samples. (d) Comparison of the mean expression profile of probe sets representing Ciita (blue) and Zbtb32 (red) across the 84 samples. (a–d) Samples are grouped according to cell type and are arranged in order of presentation as listed in Table S1. For each cell population mean expression levels are presented and the number of replicates is indicated in parenthesis on the x-axis. Red-boxed area indicates the B1 B-cell data sets. (e) Cartoon illustrating the putative functions of all the genes represented in cluster 13 (black font) and 172 (blue font) in B1 B cells. These genes were then classified into groupings of related cellular function based on published data from literature searches and bioinformatics data bases.