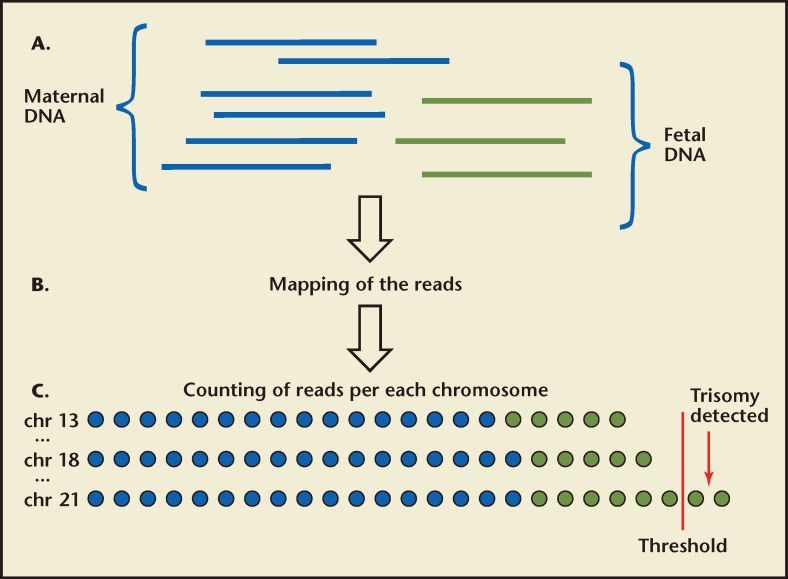
Figure 1.
Massively parallel shotgun sequencing for the noninvasive prenatal detection of fetal chromosomal aneuploidy. (A) Fetal DNA (green) circulates in the maternal plasma as a minor population among a high background of maternal DNA (blue). A sample of maternal plasma is obtained. Short fragments of cell-free DNA are then sequenced. (B) The chromosomal origin of each sequence is identified by mapping the reads to the human reference genome. (C) The number of unique sequences mapped to each chromosome is then counted and the number of sequences for the chromosomes of interest is determined in subsequent analysis. Aneuploidy is detected by various statistical techniques based on the number of reads representing the chromosome of interest compared with the other chromosomes. The blue and green circles represent the reads mapped to each chromosome from maternal and fetal DNA fractions, respectively. However, there is no way to distinguish the origin of the individual reads; the only difference is in the total number of reads that are detected. If the reads exceed that expected, the fetus is trisomic for that chromosome (shown for chr 21 above).