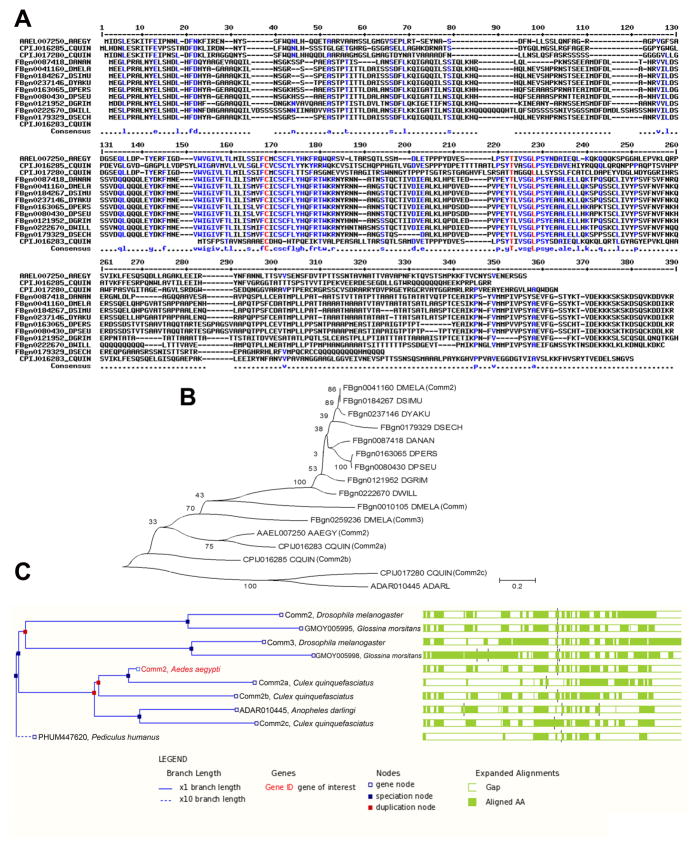
Fig. 1. Sequence analysis of insect Comm family proteins.
A. A partial multiple alignment of deduced amino acid sequences of the N-termini of predicted comm2 orthologs among mosquito and fruit fly species is shown. The gene accession numbers and species are indicated to the left of the sequences in the alignment. The amino acid position of aligned sequences is indicated at the top, while the consensus sequence of the alignment is shown below the aligned sequences. B. A neighbor-joining phylogenetic tree of C. quinquefasciatus Comm proteins along with orthologous proteins identified in A. aegypti and Drosophila species is shown. All three Comm family proteins of D. melanogaster (Comm, Comm2, and Comm3) are included in the tree. The percentage of replicate trees in which proteins clustered together through bootstrap testing is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The scale of the evolutionary distances as units of the number of amino acid substitutions per site is shown. C. The amino acid sequence conservation of selected orthologs of AAEL007250, as inferred from the GBrowse tool of VectorBase (2009), shown at right corresponds to the phylogenetic relationships among the proteins shown on the left. The percentages of sequence identities, sequence gaps and details of tree nodes are shown below the figure as captions generated by Gbrowse. Abbreviations in this figure are as follows: A. aegypti = AAEGY, C. quinquefasciatus = CQUIN, D. ananassae = DANAN, D. melanogaster = DMELA, D. simulans = DSIMU, D. yakuba = DYAKU, D. persimilis = DPERS, D. pseudoobscura = DPSEU, D. grimshawi = DGRIM, D. willistoni = DWILL, D. sechellia = DSECH.