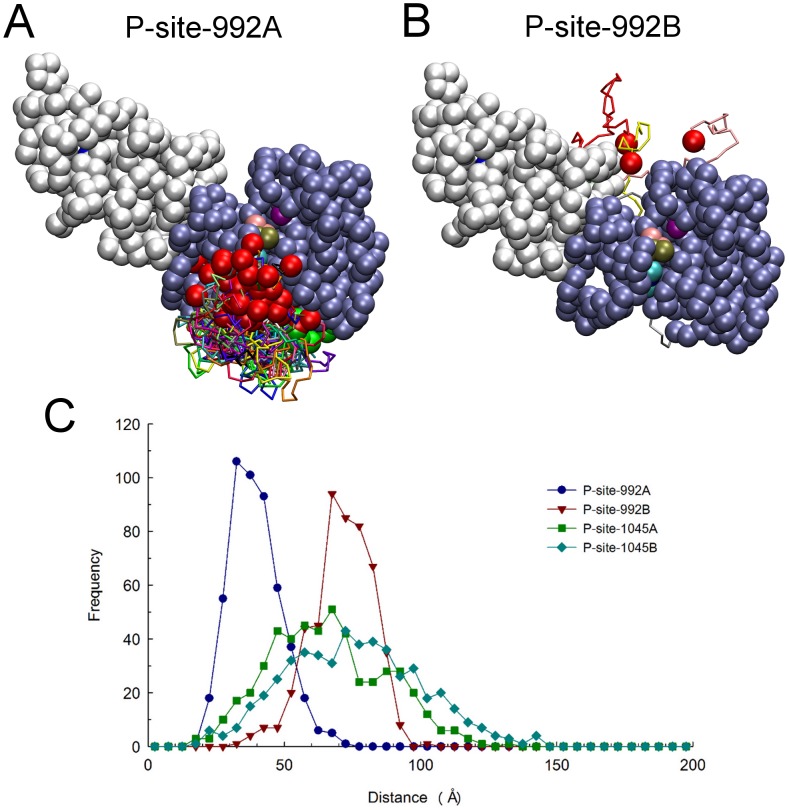
Figure 7. Relative catalytic site access of P-site-992 of the receiver and activator molecules.
Structures were examined among a set of 650 selected at 100 µsec molecular trajectories of EGFR dimer motion (from simulations in which P-site/catalytic site residues pairs were not considered to form native contacts), to identify subsets in which P-site Tyr-992 of the receiver and activator molecules were within 30 Å of catalytic Asp-813 of the receiver. Compared here are the identified catalytic site excursions of P-site-922 of the receiver (A) and activator (B) molecules, after alignment of receiver PTK domains (residues 679 to 967) in the identified structures. One receiver and one activator PTK domain are shown as ice-blue and white beads, respectively, with the catalytic Asp-813 as a purple bead and the bound nucleotide substrate complex as beads with CPK coloring. The backbones of the partial CT domains of receiver (residues 968 to 991) and activator (residues 960 to 991) molecules are shown in different colors, with residue 967 or 959 at the origin of these domains and their target Tyr-992 depicted as green and red beads, respectively. Comparison of the two panels indicates that P-site-992 of the receiver made much more frequent excursions near the catalytic site. (C) Distances from the pseudo-atoms representing tyrosine residues of the indicated P-sites to the γ-phosphate of the AMPPMP substrate bound in the catalytic site were quantified for each of the five hundred randomized structural models used as initial structures for simulations and the distance distributions plotted. Note that while there was an obvious bias in the catalytic site proximities of the receiver P-site-992 (P-site-992A) versus that of the activator (P-site-992B), this bias was markedly reduced for P-sites more distal to the kinase core (e.g. P-site-1045A versus -1045B).