Abstract
DNA damage plays a causal role in numerous disease processes. Hence, it is suggested that DNA repair proteins, which maintain the integrity of the nuclear and mitochondrial genomes, play a critical role in reducing the onset of multiple diseases, including cancer, diabetes and neurodegeneration. As the primary DNA polymerase involved in base excision repair, DNA polymerase β (Polβ) has been implicated in multiple cellular processes, including genome maintenance and telomere processing and is suggested to play a role in oncogenic transformation, cell viability following stress and the cellular response to radiation, chemotherapy and environmental genotoxicants. Therefore, Polβ inhibitors may prove to be effective in cancer treatment. However, Polβ has a complex and highly regulated role in DNA metabolism. This complicates the development of effective Polβ-specific inhibitors useful for improving chemotherapy and radiation response without impacting normal cellular function. With multiple enzymatic activities, numerous binding partners and complex modes of regulation from post-translational modifications, there are many opportunities for Polβ inhibition that have yet to be resolved. To shed light on the varying possibilities and approaches of targeting Polβ for potential therapeutic intervention, we summarize the reported small molecule inhibitors of Polβ and discuss the genetic, biochemical and chemical studies that implicate additional options for Polβ inhibition. Further, we offer suggestions on possible inhibitor combinatorial approaches and the potential for tumor specificity for Polβ-inhibitors.
Keywords: Base excision repair, chemotherapy, DNA polymerase β, lyase, polymerase, protein-protein interactions, small-molecule inhibitors
INTRODUCTION
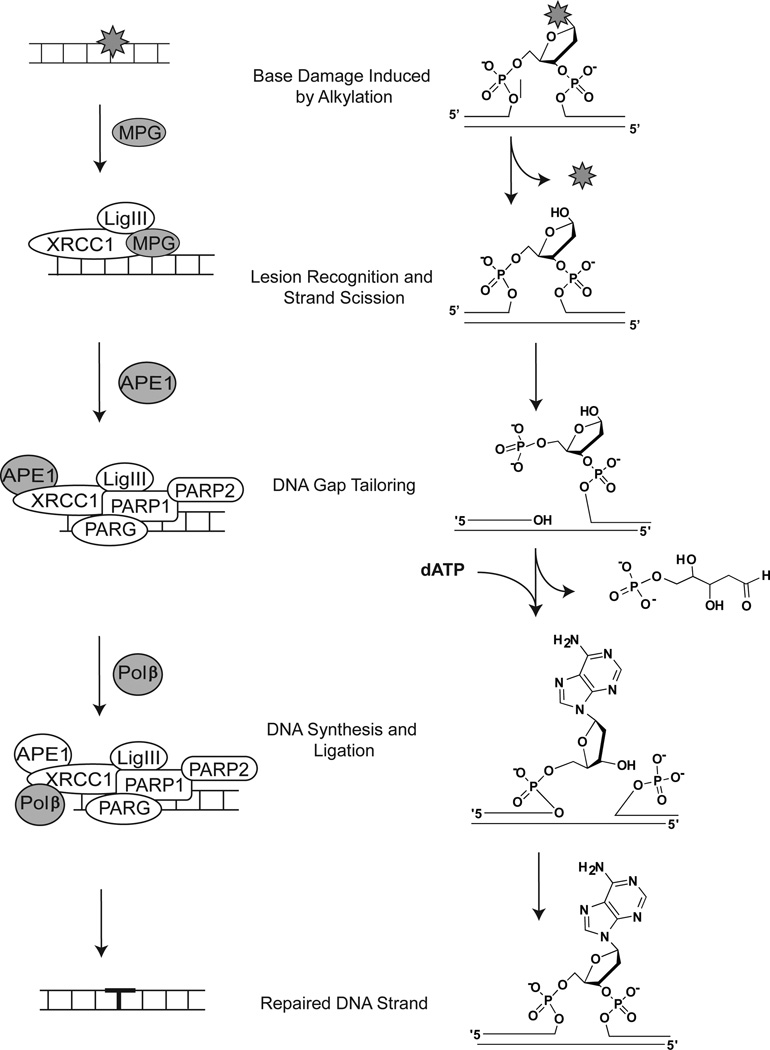
DNA polymerase β (Polβ) is a member of the X family of DNA polymerases [1, 2]. The POLB gene spans 14 exons across 33 kb and is localized to chromosome 8. A summary of genetic and physical characteristics of Polβ, along with links to several databases with additional details on Polβ, is shown in Table 1. Polβ has been implicated in several cellular functions, including genome stability [3], telomere maintenance [4–6] and meiosis [7]. Defects in Polβ have been linked with cancer [8, 9], aging [10], neurodegeneration [11, 12] and its expression is critical for the cellular response to environmental and chemotherapeutic genotoxins [13]. This latter function involves its primary role as the major DNA polymerase in the base excision repair (BER) pathway. A model for the BER proteins involved in the repair of temozolomide (TMZ)-induced lesions is depicted in Fig. (1), along with the chemical nature of each repair intermediate. In mammalian BER, a damaged base residue, such as those induced by the chemotherapeutic alkylating agent TMZ [14] is removed by a lesion-specific DNA glycosylase [15, 16]. Alkylation-induced base adducts, such as the N7-MeG and N3-MeA base lesions induced by TMZ, are removed by methylpurine (alkyladenine) DNA glycosylase. This protein has several designations, including MPG, AAG or ANPG but for clarity we will refer to it herein as MPG. The resulting abasic site is incised by apurinic/apyrimidinic endonuclease (APE1), leaving a single-nucleotide gap in the DNA strand with 3’-OH and 5’-deoxyribose phosphate (5’dRP) groups at the margins. Poly(ADP-ribose)polymerase-1 (PARP1), together with poly(ADP-ribose)polymerase-2 (PARP2) and the catabolic enzyme poly(ADP-ribose)glycohydrolase (PARG), are then suggested to be recruited to the APE1-mediated strand-break. It has been postulated that low-level activation of PARP1 and the resultant synthesis of poly(ADP-ribose) (PAR) facilitates recruitment of the downstream BER proteins XRCC1, DNA ligase IIIa (LigIIIa) and Polβ to stimulate and complete DNA repair [17].
Table 1.
Genetic and physical characteristics of Polβ*.
| Gene Name | POLB | [1] |
|---|---|---|
| NCBI Gene ID | 5423 | http://www.ncbi.nlm.nih.gov/gene/5423 |
| NCBI-gi | 4505931 | http://www.ncbi.nlm.nih.gov/protein/4505931 |
| Accession Code | NM_002690 | http://www.ncbi.nlm.nih.gov/nuccore/NM_002690 |
| GenBank | NP_002681 | http://www.ncbi.nlm.nih.gov/protein/NP_002681 |
| Uniprot | P06746 | http://www.uniprot.org/uniprot/P06746 |
| STRING | P06746 | http://string.embl.de/newstring_cgi/show_network_section.pl?identifier=P06746 |
| HPRD | 7517 | http://www.hprd.org/summary?hprd_id=07517&isoform_id=07517_1&isoform_name=isoform_1 |
| Chromosomal location | 8p11.2 | [181, 182] |
| Gene length | 33 kb | [183] |
| Protein size (MW) | 39 kDa | [3, 18] |
Additional details of Polβ and other DNA Repair genes may be found at: https://dnapittcrew.upmc.com/ and http://sciencepark.mdanderson.org/labs/wood/DNA_Repair_Genes.html
Fig. 1. Model for MPG-initiated BER.
This model depicts the proteins and chemical structures of a TMZ-induced base lesion (N3-MeA) and the corresponding BER intermediates following BER initiated by the methylpurine DNA glycosylase, MPG. The chemistry of the lesion and the repair intermediates throughout the repair process are shown on the right, highlighting the three major steps for BER: Lesion Recognition/Strand Scission, Gap Tailoring and DNA Synthesis/Ligation. The structures on the left depict the protein complexes required for completion of each step in BER initiated by MPG.
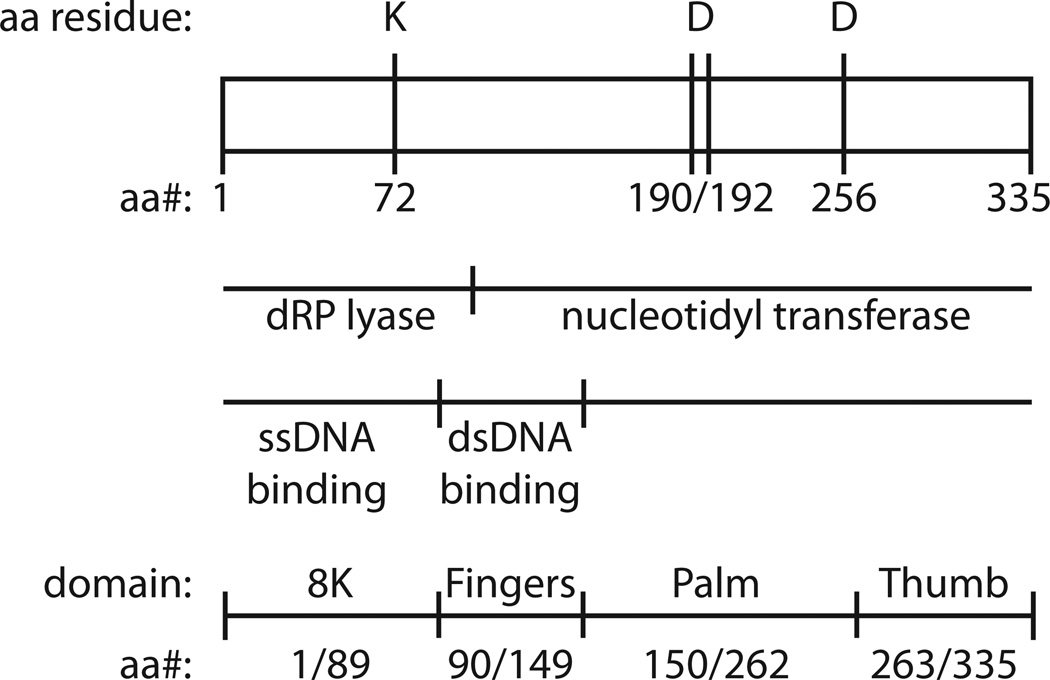
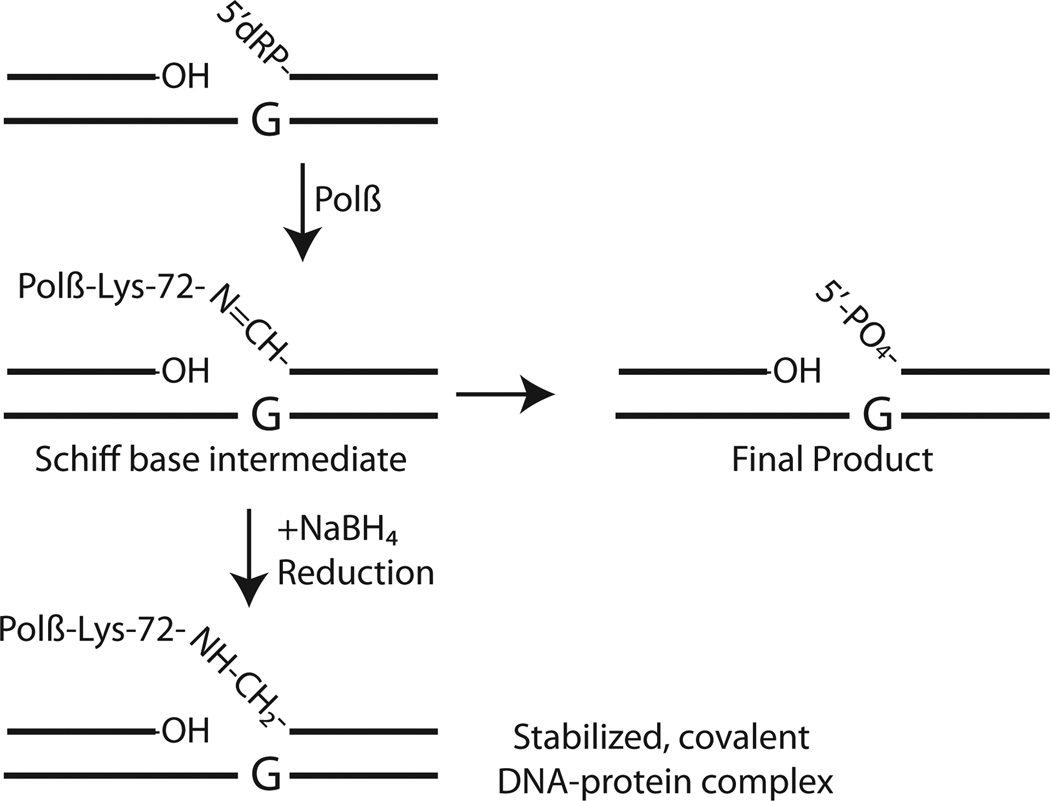
Although the POLB gene is quite large, the protein encoded by POLB is the smallest of the human DNA polymerases [3, 18]. Polβ is a bi-functional, two-domain, single-polypeptide 39kDa enzyme [18]. Structurally, Polβ is similar to other DNA polymerases in which the polymerase domain is further divided into sub-domains referred to as the fingers, palm and thumb (Fig. (2)). Detailed structural characterization of Polβ has been summarized elsewhere [18, 19]. The polymerase or nucleotidyltransferase activity, responsible for gap-filling DNA synthesis in BER, resides in the C-terminal 31kDa domain and contains three aspartic acid (D) residues (190, 192 and 256) required for activity (Fig. (2)). A second active site in the N-terminal domain of Polβ conducts the essential gap-tailoring step in BER, the removal of the 5’dRP moiety [18, 20–24] (Fig. (2)). A close examination of the molecular mechanism of the 5’dRP lyase activity of Polβ reveals the formation of a Schiff base intermediate during the reaction, as originally reported by Wilson and colleagues [25]. In this earlier report, it was demonstrated that the Polβ 5’dRP lyase reaction proceeded through an imine-intermediate, implicating one or more of 4 lysine amino acid residues (K35, K68, K72 and K84) as the 5’dRP lyase active site catalytic nucleophile [25], although later studies have defined K72 as the essential nucleophile [26]. Once the epsilon-amino group of the active site lysine has begun the nucleophilic attack on the C-1’ atom of the sugar within the sugar-phosphate BER intermediate (5’dRP), the BER reaction intermediate may be covalently trapped by reduction with sodium borohydride (NaBH4), resulting in a covalently bound substrate-enzyme complex, as depicted in Fig. (3). The 5’dRP lyase activity of Polβ is the predominant BER-dependent 5’dRP lyase activity observed in mammalian cells for the repair of alkylated [27] and oxidized [28] bases. However, biochemical and genetic analyses have identified additional proteins with 5’dRP lyase activities that may contribute to gap tailoring in nuclear and mitochondrial BER, including DNA polymerases theta (θ), lambda (λ,), gamma (γ) and iota (ι) [24, 29–33]. Additional enzymes with similar 5’dRP lyase activities include the catalytic subunit of the HSV-1 DNA polymerase [34–36], the S. cerevisiae Trf4 protein [37], NEIL family glycosylases [38, 39], DNA polymerase β from Leishmania infantum [40], Ku [41], DNA polymerase X from African swine fever virus [42] and Deinococcus radiodurans [43], mitochondrial DNA polymerase β from Crithidia fasciculate [44], the RecJ and Fpg proteins of E. coli [45, 46], HMGA2 [47] and HMGB1 [48].
Fig. 2. DNA Polymerase β.
A linear depiction of the amino acid residues (335) of DNA Polymerase β: indicating the functional domains (dRP lyase and nucleotidyltransferase), the essential active site residues (K72 and D190/192/256), regions involved in ssDNA and dsDNA binding and the structural sub-domains as determined by crystallographic analysis (8K, fingers, palm and thumb).
Fig. 3. Schiff base formation and the 5’dRP lyase activity of Polβ.
Schematic demonstrating the Schiff base formed as an intermediate in the 5’dRP lyase reaction conducted by Polβ, mediated by the nucleophile, K72. Completion of 5’dRP removal yields a final product (right side) that is further processed by Polβ by addition of a single nucleotide and ligation mediated by the XRCC1/LigIII heterodimer (see Fig. (1)). Evidence of the Schiff base is demonstrated by the stabilized DNA-protein complex formed upon reduction with NaBH4 (bottom left).
Polβ has two active sites and several critical functional domains that may be considered as targets to inhibit Polβ and BER. In addition, Polβ-dependent BER is a highly complex process involving several proteins and requires multiple protein-protein interactions for repair (depending on the base lesion) [3], suggesting that interference with DNA binding or with the formation of one or more essential protein-protein interactions may in effect inhibit Polβ-mediated repair. Further, cancer mutations or cancer specific truncations of Polβ offer additional opportunities for BER-specific modulation and recently, it has been suggested by us and others that inhibition of Polβ may provide an opportunity for synthetic lethality associated with cancer specific genotypes. BER and Polβ can also be regulated by protein modifications but an understanding of this aspect of BER and Polβ regulation is in its infancy [3, 49]. For example, Polβ can be modified by acetyl, methyl and phosphate groups, yet little is known about the functional impact of these modifications [50, 51]. More recently, it has been suggested that Polβ is modified by ubiquitin, leading to proteasome-mediated degradation [52–54]. As will be discussed throughout this review, the central and pivotal role of Polβ in BER implicates this protein as a prime target to enhance the response to chemotherapeutic agents or radiation. The complexity of BER and the ubiquitous role of Polβ in cellular metabolism suggest that the design and effectiveness of a robust Polβ inhibitor will be a significantly complex but rewarding challenge.
INHIBITING POLβ-DEPENDENT DNA SYNTHESIS
Initial attempts to regulate cancer cell growth focused on inhibiting DNA synthesis in order to regulate the uncontrolled replication of the cancer cell [55, 56]. Such inhibition can be accomplished directly or indirectly [55]. Indirect methods include damaging the DNA sufficiently to prevent replication [57] or depletion of nucleotide pools with anti-metabolites such as methotrexate [58]. Alternatively, components of the DNA replication machinery can be inhibited directly. This idea has led to numerous inhibitors of DNA polymerases useful in the treatment of diseases associated with cellular hyperproliferation, including cancer and viral infection [55, 56]. However, the requirement for normal cellular replication severely limits the utility of general DNA polymerase inhibitors. The identification of 15 mammalian DNA polymerases [1, 59] involved in nuclear and mitochondrial DNA replication, repair, recombination and translesion DNA synthesis complicates the utility of DNA polymerase inhibitors, requiring a high degree of selectivity and specificity. However, the unique role of repair and translesion DNA polymerases, especially in response to radiation and chemotherapy [56, 60, 61] suggests an opportunity for selectivity and specificity of DNA polymerases in response to some anti-cancer agents. The unique role of Polβ in BER, in particular in response to chemotherapy [60, 62], suggests that targeting the DNA synthesis activity of Polβ may offer a selective advantage in the treatment of cancer. A discussion on the role of Polβ in translesion DNA synthesis is presented by Sharma and colleagues elsewhere in this Special Edition and will not be covered here. Below, we have summarized past and present advances in the development of small molecule inhibitors specific to the DNA polymerase activity of Polβ.
Natural Products as Inhibitors of Polβ-Mediated DNA Synthesis
Hecht and colleagues utilized a standardized approach of ‘bioassay-guided fractionization’ to identify a series of natural-product-derived Polβ inhibitors isolated from crude plant extracts (e.g., aqueous or methyl ethyl ketone extracts) [63–70]. The assay used was a standard in vitro DNA polymerase assay using either purified calf thymus Polβ or recombinant rat or human Polβ, in the presence or absence of bovine serum albumen (BSA), monitoring the incorporation of [3H]dTTP in response to the presence or absence of the inhibitor or extract. In some cases, the compounds were resynthesized to validate the results.
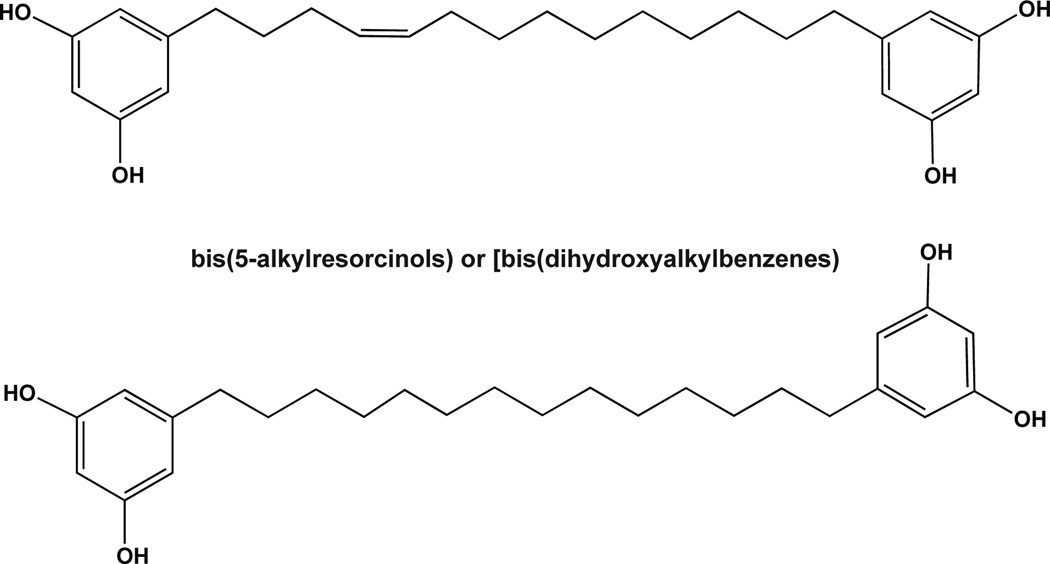
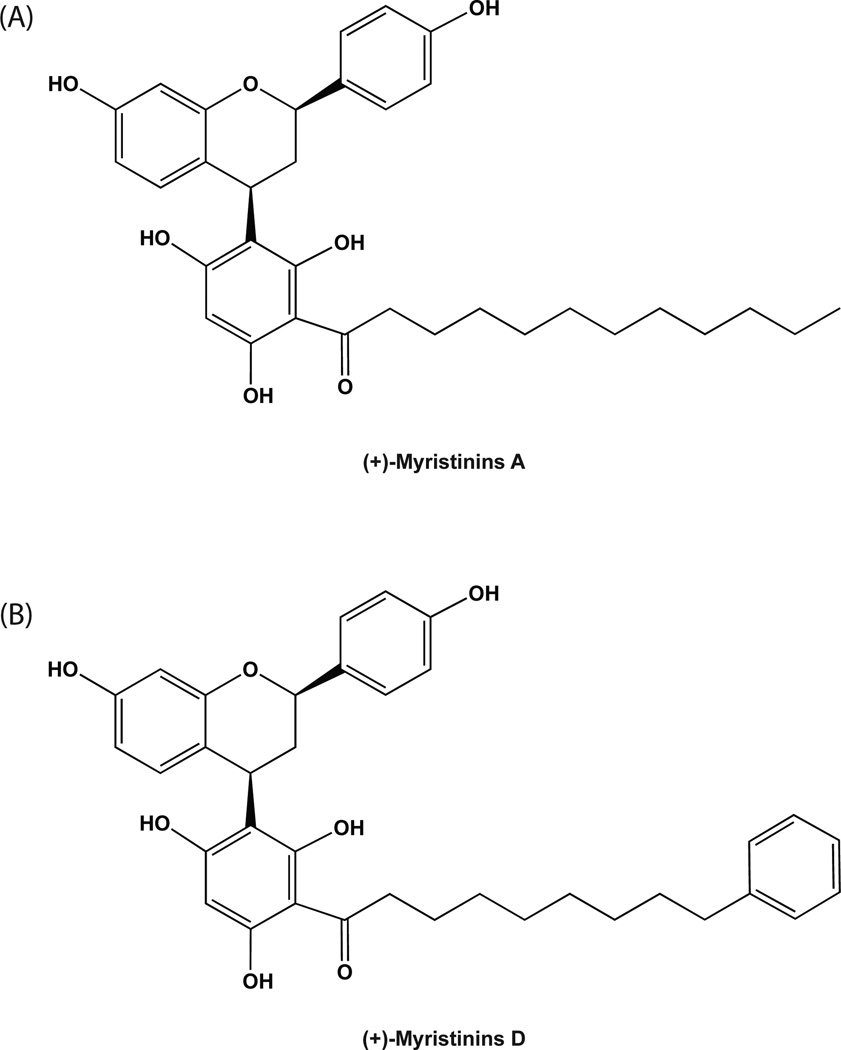
The earliest studies yielded the identification of several alkylresorcinol analogs [65, 69], including one previously identified as bis-5-alkylresorcinol [65]. In addition, two novel alkylresorcinol analogs were identified, depicted in Fig. (4). The most potent of these exhibited IC50 values as low as 5.3 µM [65, 69]. However, specificity and selectivity of these compounds has yet to be evaluated. Interestingly, these two alkylresorcinol analogs, as shown in Fig. (4), initiate Cu2+-dependent DNA cleavage, prompting Hecht and colleagues to evaluate additional classes of compounds that can both inhibit Polβ and cleave DNA. Recently, Hecht and colleagues isolated a set of flavinoids [(+)-myristinin A and D], shown in Fig. (5), from knema elegans [71] and later they accomplished the complete chemical synthesis of (+)-myristinin A [72]. These also have potent Polβ inhibitory activity and mediate Cu2+-dependent DNA cleavage. When evaluated for an impact on cellular viability at 10 µM, each appeared to be as effective as 0.075µM bleomycin. The combination of bleomycin plus the alkylresorcinol compound or the flavinoids yielded either a similar impact or slightly greater impact on cell growth and viability. It was therefore suggested that these alkylresorcinol and flavinoid compounds damage DNA and then inhibit the repair of the induced DNA damage. It remains to be determined if the damage induced by either the alkylresorcinol or flavinoid compounds is repaired by Polβ.
Fig. 4. Novel alkylresorcinol inhibitors of Polβ.
Schematic depicting two novel alkylresorcinol analogs found to inhibit Polβ.
Fig. 5. Flavinoid inhibitors of Polβ.
Structure of two flavinoids isolated from Knema elegans and identified as inhibitors of Polβ. The parent structure is shown, with varying R groups, depicting (A)(+)-myristinin A and (B)(+)-myristinin D.
The same group also reported the isolation of several triterpenoids and similar multicyclic compounds isolated from several plant species that can inhibit DNA synthesis mediated by Polβ. Compounds were isolated from Brackenridgea nitida [68], Bleasdalea bleasdalei [68], Freziera sp. [66], Baeckea gunniana [64], Tetracera boiviniana [63] and Sandoricum koetjape [70]. None yielded compounds with IC50 values lower than 2 µM. More recently, Cazaux and colleagues utilized a high-throughput approach, screening over 8,000 natural products (extracts) to identify an inhibitor of Polβ-mediated DNA synthesis. In this screen, they identified masticadienonic acid (MA), demonstrating a small degree of specificity as MA does not appear to inhibit the enzyme activity of Polδ. Since MA enhanced the cell killing effect of cisplatin [73], it was suggested that MA may affect the translesion-synthesis role of Polβ (See article by Sharma and colleagues elsewhere in this Special Edition). Other natural products found to inhibit Polβ include fatty acid derivatives [74], azaphilones isolated from cultures of Talaromyces sp. [75] and 1-deoxyrubralactone [76], among others we may have inadvertently missed. It remains to be established if any of these are selective or specific to Polβ, as compared to other Pol X family members or other DNA polymerases.
Sulfolipids
Using a similar functional screening paradigm, Sakaguchi and colleagues have identified several novel classes of compounds that inhibit Polβ. By analyzing extracts of the pteridophyte A. niponicum, they isolated a series of sulfolipids with inhibitory activity towards Polα and Polβ [77], with IC50 values of 6 and 8 µM, respectively. This discovery was followed by several reports describing a series of sulfolipids of the sulfo-quinovosyl-acyl-glycerol (SQAG) class [78–81]. All seem to function as competitive inhibitors of Polβ (with respect to template/primer or dNTP) whereas some exhibit non-competitive inhibition of Polα. As with Lithocholic acid (LCA), MA and related fatty acid or steroid-based lipid derivatives (see below), the sulfolipids appear to bind to the N-terminal 8kDa domain and interfere with DNA binding, preventing nucleotidyltransferase (polymerase) activity [82]. Since many of these sulfolipids contain an esterified form of fatty acids similar to those known to inhibit Polβ (see below), it is speculated that both fatty acids and sulfolipids may inhibit Polβ via a similar mechanism [78, 80]. However, it is noted that both the sulfate in the quinovose moiety as well as the fatty acid component of these sulfolipid compounds are critical for inhibition of Polβ-mediated nucleotidyltransferase activity [78–80]. Similarly, it was established that sulfated glycoglycerolipids (e.g., KN-208) are effective in inhibiting Polβ (Ki = 0.05 µM), yet as with the others mentioned here, this compound also inhibits Polα although at a 10-fold higher concentration [83].
Interestingly, many of these sulfolipids have additional cellular effects that may need to be considered if these are to be developed clinically. For example, Sulfo-quinovosylmonoacyl-glycerol (SQMG), also shown to inhibit Polβ [82], triggers cell cycle arrest [84] and has anti-angiogenic effects, likely via down-regulation of Tie2 [85]. Further, SQMG functions as a radiosensitizer [86], although it has not been determined if any of the radiosensitizing effect is via inhibition of Polβ or other DNA polymerases.
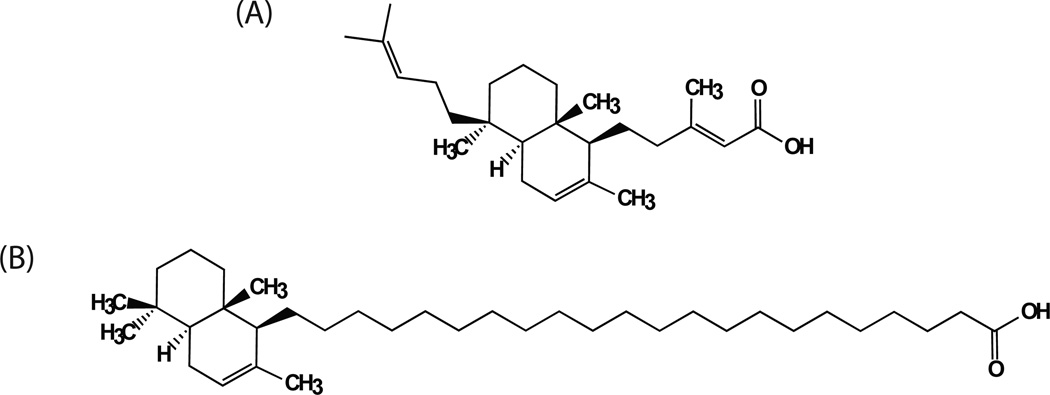
KA-A
Kohamaic acid A (KA-A) is a sesterterpenic acid that was isolated from Ircinia sp., a marine sponge, and was first reported to inhibit cell division of fertilized sea urchin eggs [87]. The effect on cell division was suggested to proceed via a block or inhibition of sea urchin replication-essential DNA polymerases. More detailed analysis of KA-A and a series of analogs [88, 89] revealed broad spectrum inhibition of human DNA polymerases α, β, δ and γ, suggesting a possible universal mechanism of inhibition of DNA polymerases but a clear lack of specificity for Polβ. The structure of the natural compound KA-A and the most effective derivative [Compound #11; (1S*, 4aS*, 8aS*)−17-(1,4,4a,5,6,7,8,8a–octahydro-2,5,5,8a–tetramethylnaphthalen-1-yl)heptanoic acid] [88] is shown in Fig. (6). Compound #11 acts as a competitive inhibitor of Polβ with Ki values of 1.9 µM (template/primer) and 2.3 µM (nucleotide). Molecular modelling studies suggest KA-A derivative #11 binds to the 8kDa domain of Polβ along the interface that interacts with the ssDNA of a template/primer. Such inhibition would be similar to that of other fatty acid derivatives [90].
Fig. 6. Kohamaic acid A and a chemically derived fatty acid derivative.
The structure of (A) the Ircinia sp. derived compound Kohamaic acid A (KA-A) and (B) the fatty acid derivative Compound #11; [1S*,4aS*,8aS*)-17-(1,4,4a,5,6,7,8,8a–octahydro-2,5,5,8a–tetramethylnaphthalen-1-yl)heptanoic acid] [88].
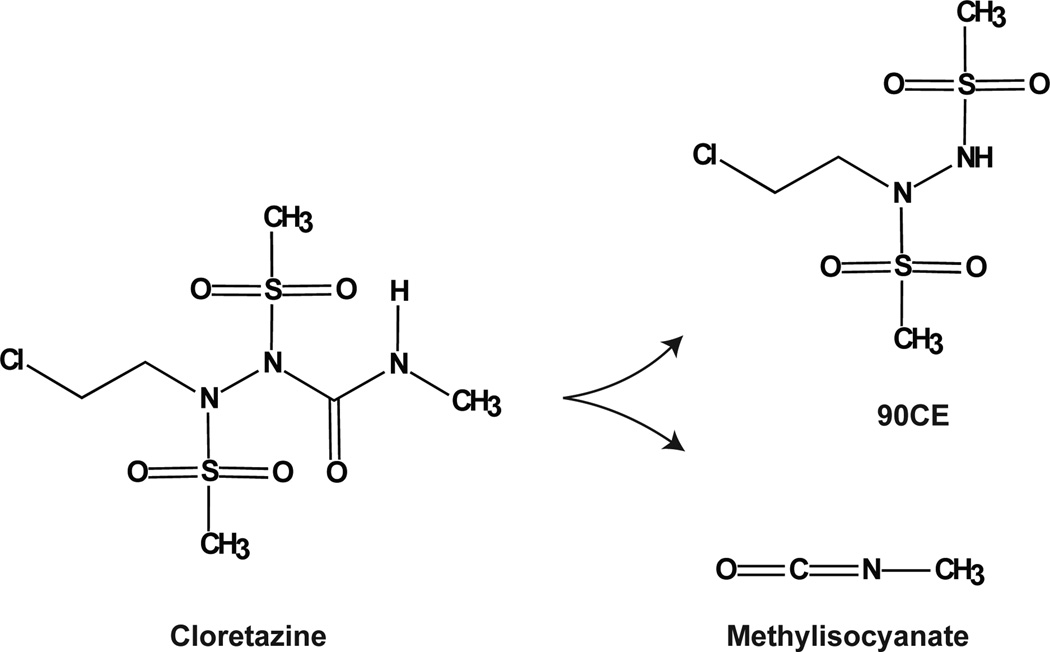
Cloretazine
The nucleotidyltransferase activity of Polβ has also been reported to be inhibited by the inter-strand DNA crosslinking agent Cloretazine [91] or 1,2-Bis(methylsulfonyl)-1-(2-chloroethyl)-2-[(2-methylamino)carbonyl] hydrazine. This agent, also referred to as Laromustine or VNP-40101M [92] is currently in several stages of clinical evaluation [93–95]. Cloretazine is activated under aqueous conditions, yielding the two reactive molecules 90CE and methylisocyanate, as shown in Fig. (7). The 90CE derivative is responsible for alkylation of DNA at the O6 position of guanine bases, triggering cytotoxicity resulting from the formation of interstrand cross-links to cytosine. However, cytotoxicity is alleviated in cells expressing the direct-reversal repair protein O6-methylguanine-DNA methyltransferase (MGMT) [96]. The methylisocyanate compound induces carbamoylation of protein sulfhydryl groups [97]. Rice and colleagues observed that the Cloretazine derivative 90CE has no effect on Polβ whereas the analog 101MDCE, which retains the carbamoylating activity, has an IC50 value of 92 µM, suggesting Cloretazine inhibits Polβ by methylisocyanate-mediated carbamoylation of an active-site sulfhydryl group, attenuating DNA polymerase activity. We await validation of this initial report.
Fig. 7. Cloretazine and reactive metabolites.
Shown is the structure of Cloretazine and the reactive breakdown products 90CE and methylisocyanate [91, 96].
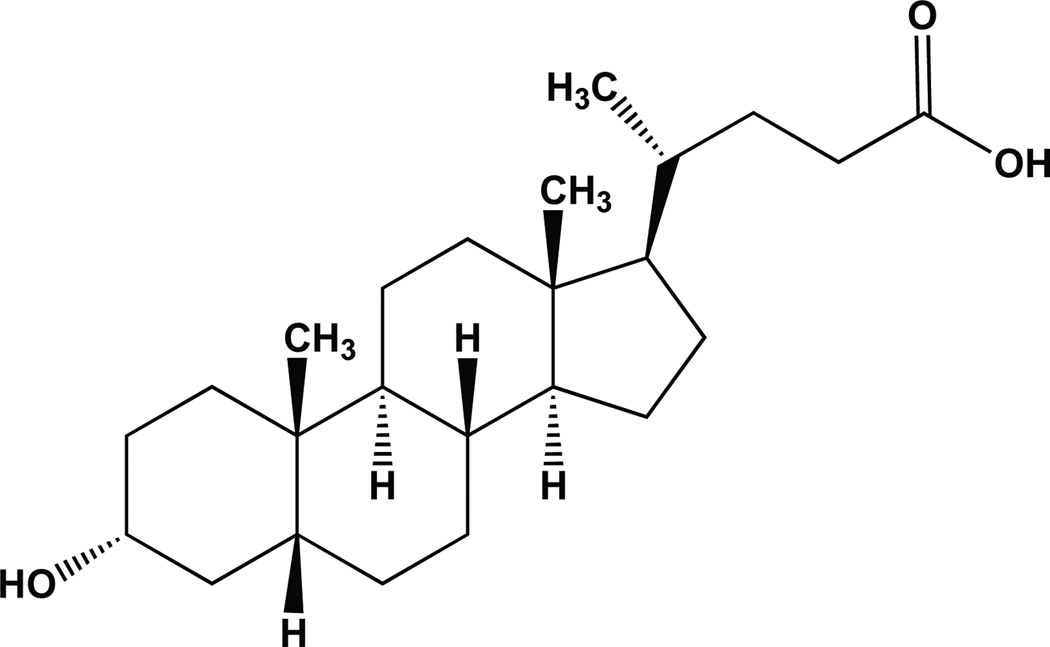
Lithocholic Acid
One small molecule that has received considerable attention as a purported specific inhibitor of Polβ is Lithocholic acid (LCA) − see Fig. (8). Originally identified by DeClercq and colleagues as a cholic acid derivative with selectivity for HIV-1-mediated viral replication [98], LCA was subsequently shown to promote N-methyl-N'-nitro-N-nitrosoguanidine (MNNG)-mediated tumor growth [99–101]. Since MNNG induces DNA damage that is repaired by Polβ and the BER pathway [13], Yoshida and colleagues then hypothesized that the tumor promoting action of LCA may be mediated by inhibiting Polβ and preventing repair of the MNNG-induced DNA damage. After evaluating seventeen related bile acids, it was determined that LCA and select derivatives were potent inhibitors of Polβ. Note however, that LCA was also found to inhibit Polα, Polδ, Polε and Polγ although Polβ was reported as the most sensitive [99]. In follow-up analyses by Sakaguchi and colleagues, it was determined that LCA bound to the N-terminal 8kDa domain of Polβ [102]. Although the binding affinity of LCA to Polβ was sufficient to allow modified forms of LCA (e.g., biotinylated) for use in the purification and affinity capture of Polβ [103, 104], this approach can also be used to purify Polα [103, 104] and LCA also binds tightly to DNA Topoisomerase II [105], suggesting some level of off-target effects of LCA with regard to Polβ.
Fig. 8. Lithocholic acid.
Structure of Lithocholic acid: (4R)-4-[(3R,5R,8R,9S,10S,13R,14S, 17R)-3-hydroxy-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H–cyclopenta[a]phenanthren-17-yl]pentanoic acid.
Recently, LCA was shown to potentiate the cell killing effect of TMZ, a clinical alkylating agent previously shown to induce BER specific DNA damage requiring Polβ for cell survival [14, 60, 62, 106]. Importantly, it was shown that potentiation of LCA on TMZ was enhanced in cells with a null mutation in the BRCA2 gene [107]. In this latest report, Glazer, Sweasy and colleagues very clearly demonstrate that LCA inhibits both the nucleotidyltransferase and the 5’dRP lyase activity of purified Polβ, as well as inhibiting Polβ in a complete BER reaction. Under the assumption that LCA is specific for Polβ, this recent study was then extended to cells deficient or proficient for BRCA2, showing synergy between LCA and TMZ. However, these studies failed to effectively demonstrate that the synergy between LCA and TMZ is the result of specific or selective Polβ inhibition. For example, LCA treatment yields a similar level of synergy with TMZ in both wild-type (WT) and MPG(AAG) knockout (KO) cells [107]. Since it was earlier demonstrated that Polβ/AAG double-KO cells are resistant to the cell killing effects of alkylating agents (as compared to Polβ KO cells) [108], the reported synergy of LCA and TMZ in AAG KO cells [107] likely involves at least some level of cell killing resulting from either non-specific effects of LCA or inhibition unrelated to Polβ. As with many of the compounds discussed herein, further tests are required to clearly demonstrate if LCA is selective or specific to Polβ in human cells, especially given the previous evidence for inhibition of other human DNA polymerases [99]. Further, LCA has additional cellular effects, likely both related and unrelated to its function as a ligand for the Vitamin D receptor, including regulation of NF- κB and Vitamin D [109, 110] as well as induction of the urokinase-type plasminogen activator receptor (uPAR) [111]. Most recently, LCA was identified in a large-scale chemical and genetic screen to extend yeast chronological life span [112], an effect clearly not mediated through Polβ.
DNA BINDING INHIBITION
NSC666715
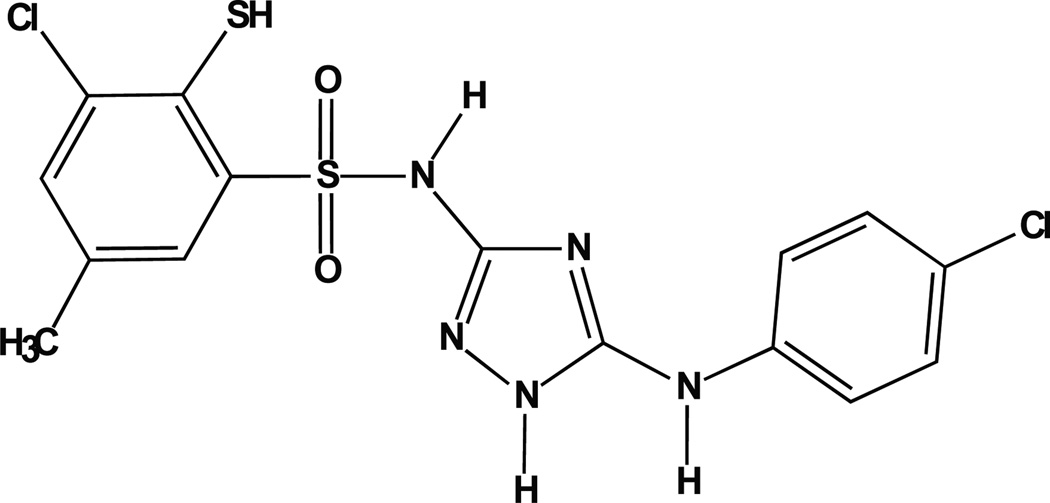
As depicted in Fig. (2), Polβ has two essential DNA binding domains [18]. The 8kDa domain, corresponding to amino acid residues 1–89, binds to ssDNA [18] with a preference for a small gap with a 5’phosphate [23]. The fingers sub-domain, corresponding to amino acid residues 90–149, encodes the dsDNA-binding domain [18]. However, there is likely some level of cooperative binding since the C-terminal 31kDa Polβ fragment, corresponding to amino acid residues 90–335, has minimal DNA binding and DNA polymerase activity [27]. Given the extraordinary number of proteins capable of binding DNA plus the potential for a high degree of similarity among proteins that bind DNA in a non-sequence dependent manner, it was surprising that a relatively specific Polβ inhibitor was recently reported that appears to function precisely by inhibiting the binding of Polβ to the damaged DNA under repair [113]. In this report, Narayan and colleagues reported promising results with the compound NSC666715 [113], depicted in Fig. (9). This compound (NSC666715; IUPAC designation = 4-chloro-N-[5-(4-chloroanilino)-1H-1,2,4-triazol-3-yl]-5-methyl-2-sulfa-nylbenzene-sulfonamide) was first synthesized as an anti-HIV compound, designed for inhibition of viral integration [114–116]. Narayan and colleagues used a molecular docking approach to identify small molecules that would bind to a recently identified APC-binding site on Polβ [117, 118]. Using purified proteins, they find that NSC666715 inhibits both the 5’dRP lyase and DNA synthesis steps catalyzed by Polβ, resulting in an overall block to BER. Treatment with NSC666715 was effective in enhancing the efficacy of TMZ in a mouse xenograft model using the HCT-116 colon tumor model [113]. In this report, the data suggests that NSC666715 prevents Polβ from binding damaged DNA. The specificity and selectivity of NSC666715 remains to be determined, as treatment with NSC666715 alone resulted in the inhibition of tumor cell growth in the absence of TMZ, an effect not likely to be the result of Polβ inhibition. Regardless, this initial report is a promising example of targeting Polβ by preventing the ability to recognize and bind to damaged DNA.
Fig. 9. Structure of NSC666715.
Schematic depicting the chemical structure of 4-chloro-N-[5-(4-chloroanilino)-1H-1,2,4-triazol-3-yl]-5-methyl–2-sulfanylbenzenesulfonamide (NSC666715), initially reported as a potential inhibitor of HIV integrase [114–116] and later identified as an inhibitor of Polβ [113].
DISRUPTING POLβ-DEPENDENT DNA REPAIR COMPLEXES
As our understanding of the BER pathway has evolved, especially the molecular details of each step in the mechanism of BER, it has become clear that repair of base lesions and related BER substrates is facilitated via a series of protein complexes that assemble at the site of the DNA lesion [3, 18, 49, 119]. This should not be surprising as essentially all proteins either function by or are regulated by specific protein-protein interactions. In fact, protein-binding interfaces may offer a greater number of targets for the development of highly specific and selective drugs. For example, one recent accounting of the number of protein-protein interactions in human cells suggest as many as 150,000 protein interaction pairs [120] and this number may be greatly enhanced when considering the increase in the number of protein products that result from alternative splicing events [121]. Further, as with other repairosomes or transient DNA Repair complexes, post-translational modification (PTM) of BER proteins impacts both function and repair complex formation [49].
Targeting the Polβ-XRCC1 Interface
A recent example of targeted disruption of a proteinprotein interaction is the disruption of the c-Myc/Max heterodimer. The oncoprotein c-Myc is over-expressed in many cancers and forms a heterodimer with Max to function as a transcription factor [122]. It has been demonstrated that small molecules can disrupt the c-Myc/Max dimer, eliminating the oncogenic potential of c-Myc overexpression [123, 124]. Similar protein-protein disrupting molecules or peptide mimetics have also been developed to target SMAC in an attempt to induce apoptosis [125, 126] although other targets have been proposed [127, 128]. In this same vein, we propose that given the number of specific protein-protein interactions that are formed during the repair of base lesions by Polβ and the BER pathway, it might also be possible to develop selective and specific BER inhibitors by targeting specific protein-protein interfaces that arise during repair. As summarized in Table 2, there are a number of proteins that functionally interact with Polβ or modulate Polβ function. These proteins have been reported to either modify Polβ (e.g., Mule) or participate in BER (e.g., LigI). However, for many of these proteins, the functional significance of binding to Polβ is unknown (e.g., TAF1D). For those that form complexes with Polβ to facilitate BER (e.g., XRCC1), we propose that targeted disruption of this complex or protein-protein interaction may lead to inhibition of Polβ-mediated BER and hence the Polβ/XRCC1 interface may provide a unique drug development target. Polβ forms a tight complex with XRCC1 [129], amenable to both NMR analysis [130, 131] and crystallographic analysis [132]. The region of interaction with Polβ is located within residues 84–183 in the N-terminal half of the XRCC1 protein [133]. Conversely, the Polβ domain that binds to XRCC1 is the C-terminal or thumb domain (see Fig. (2)). As was demonstrated using NMR spectroscopy, Polβ mutants with a L301R/V303R/V306R triple point mutation cannot interact with XRCC1 [131, 134]. These amino acid residues form part of a protusion (referred to as the V303 hairpin) that appears to insert within the Polβ/XRCC1 binding interface, as determined by crystallographic analysis [132] – see Fig. (10). Interestingly, the Polβ/XRCC1 binding interface appears to be regulated by oxidation [132]. Given the significance of the Polβ/XRCC1 interaction in BER [135–137], it is therefore likely that disruption or inhibition of BER may be accomplished by small molecules or peptide mimetics targeted to this location within the Polβ/XRCC1 binding interface.
Table 2.
DNA polymerase β interacting proteins.
| Protein abbreviation |
Protein name | Polβ interaction domain |
Method* | Functional impact | Citation |
|---|---|---|---|---|---|
| AAG (MPG) | N-methylpurine DNA glycosylase | - | IP | - | [184] |
| APC | Adenomatous polyposis coli | 8K domain (Thr79, Lys81, and Arg83) | FW, Y2H | Inhibition of Polβ lyase activity, inhibition of Polβ strand displacement synthesis | [118, 185, 186] |
| APE1 | DNA-(apurinic/apyrimidinic site) lyase | - | Y2H | Stimulation of the 5’dRP lyase activity | [187] |
| CHIP | E3 ubiquitin-protein ligase CHIP | 8K domain | - | Polyubiquitylation of Polβ | [54] |
| FEN1 | Flap structure-specific endonuclease 1 | - | IP | Stabilization of FEN1 binding to DNA, stimulation of strand displacement and flap hydrolysis | [188, 189] |
| HMGB1 | High-mobility group box 1 | - | IP | - | [48] |
| HSP27 | Heat shock protein 27 | - | IP | Thermoprotection of Polβ | [190] |
| HSP70 | Heat shock protein 70 | - | IP | Stimulation of Polβ activity, thermoprotection of Polβ | [190, 191] |
| Lig1 | DNA ligase 1 | - | IP, PD | Inhibition of strand displacement synthesis | [192] |
| MGC5306 (JOSD3; TAF1D) | TATA box-binding protein-associated factor RNA polymerase I subunit D | - | Y2H, IP | - | [193] |
| Mule | Mcl-1 ubiquitin ligase | 8K domain | - | Monoubiquitylation of Polβ | [52] |
| MYST1 | MYST histone acetyl transferase 1 | - | Y2H | - | [194] |
| NEIL1 | Nei endonuclease VIII-like 1 (E. coli) | Amino acids 1–140 | FW | NEIL1 initiated APE-1 independent BER | [195] |
| NEIL2 | Nei endonuclease VIII-like 2 (E. coli) | Amino acids 1–140 | IP, FW | NEIL2 initiated APE-1 independent BER | [196] |
| OGG1 | 8-oxoguanine DNA glycosylase 1 | - | IP | - | [184] |
| p300 | Histone acetyltransferase p300 | 8K domain | IP, PD | Acetylation of Polβ | [51] |
| p53 | Cellular tumor antigen p53 | - | FW, IP | Enhanced stabilization of Polβ to AP site DNA | [197] |
| PARP1 | Poly(ADP-ribose) polymerase 1 | Amino acids 124–335 | PD | Stimulation of Polβ strand displacement synthesis activity, enhanced BER activity | [198] |
| PARP2 | Poly(ADP-ribose) polymerase 2 | - | PD | Enhanced BER activity | [199] |
| PCNA | Proliferating cell nuclear antigen | 31K domain (Palm) | IP, Y2H | - | [200] |
| PNKP | Polynucleotide kinase 3’- phosphotase | - | IP | Enhanced single strand break repair | [201] |
| PRMT1 | Protein arginine methyltransferase 1 | 8K domain | IP, PD | Arginine methylation of Polβ | [202] |
| PRMT6 | Protein arginine methyltransferase 6 | 8K domain | IP, PD | Arginine methylation of Polβ | [203] |
| Rad9/Rad1/Hus1 (9-1-1 complex) | RAD9 homolog/RAD1 homolog/HUS1 checkpoint homolog | - | IP | Stimulation of Polβ strand displacement synthesis activity, increased Polβ affinity for 3’OH primer end | [204] |
| RPA | Replication protein A | - | PD | Reduction of Polβ gap filling efficiency opposite of 8-oxo-G | [205] |
| TLE1 | Transducin-like enhancer of split 1 | - | Y2H | - | [194] |
| Trf2 | Telomeric repeat binding factor 2 | 31K domain | Stimulation of primer extension, stimulation of strand displacement synthesis | [5, 6] | |
| WRN | Werner syndrome, RecQ helicase | - | IP, PD | Stimulation of Polβ strand displacement synthesis, unwinding of Polβ single strand intermediates | [206] |
| XRCC1 | X-ray repair complementing defective repair in Chinese hamster cells | 31K domain (thumb-palm domains) | FW, Y2H, PD | Scaffolding of BER complex, inhibition of strand displacement | [133] |
IP, immunoprecipitation; PD, GST- or other tagged protein pulldown; Y2H, Yeast two-hybrid; FW, Far-Western.
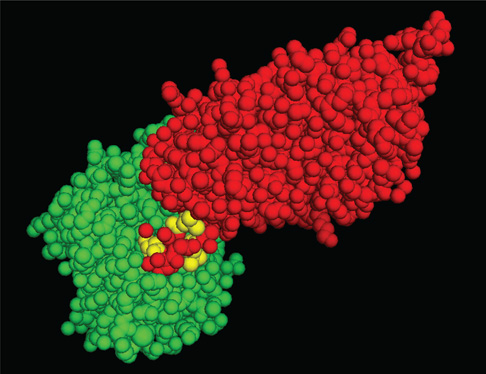
Fig. 10. Model depicting the Polβ/XRCC1 interaction.
Structure of a complex formed by the thumb sub-domain of Polβ and the N-terminal domain of XRCC1 [132]. The image is a spacefilling rendition of the amino acid residues of XRCC1 in green and the thumb sub-domain of Polβ in red. The residues in the V303 hairpin, corresponding to L301, V303 and V306 are shown in yellow. The image was generated using PyMOL (Molecular Graphics System, Version 1.2r3pre; Schrödinger, LLC; http://pymol.org/).
Modulating POLβ Function By Regulating PTMs
As discussed previously [3, 49] and mentioned above, Polβ helps facilitate BER via the formation of transient protein complexes at the site of the DNA lesion. Further, it appears that, as with many proteins, post-translational modification (PTM) of BER proteins can either enhance or disrupt BER, in some cases by modulating protein complex formation [49]. Like many BER proteins, Polβ is modified at multiple amino acid residues – summarized in Table 3. For example, Dianov and colleagues have suggested that Polβ levels are kept in check by ubiquitylation and subsequent proteosome-mediated degradation [52, 54]. Therefore, it is conceivable that a targeted approach of Polβ ubiquitylation and degradation might be feasible. Alternatively, enhancing the acetylation of K72 [51] would inhibit the 5’dRP lyase activity of Polβ. In both cases, the result would be a targeted disruption of BER. However, we concede that selective drugs to facilitate enhanced modification of Polβ is highly speculative at this time, especially considering that many of the reported Polβ-specific PTMs (Table 3) have not been validated in multiple systems and little is known regarding the functional impact of these PTMs. It is possible that many of the PTMs reported to date are not universal and/or do not impact Polβ function or Polβ-mediated BER complex formation. In addition, the protein modifiers (e.g., CHIP, PKC, p300) have multiple targets unrelated to Polβ and BER and therefore inhibition of these enzymes is not likely to be Polβ specific. We look forward to future studies in this regard.
Table 3.
Post-translational modifications of Polβ.
| Modification | Amino acids modified | Modified by | Functional impact | Citation |
|---|---|---|---|---|
| Phosphorylation | Ser 44, Ser 55 | PKC alpha | Inhibition of polymerase activity | [207] |
| Phosphorylation | Tyr 250 | n.d. | n.d. | [208, 209] |
| Acetylation | Lys 72 | p300 | Inhibition of 5’dRP lyase activity | [51] |
| Methylation | Arg 137 | PRMT1 | Inhibition of binding to PCNA | [202] |
| Methylation | Arg 83, Arg152 | PRMT6 | Stimulation of DNA binding and polymerase activity | [203] |
| Polyubiquitylation | n.d. | E3 ligase CHIP | Degradation of Polβ not involved in BER complex | [54] |
| Monoubiquitylation | Lys 41, Lys 61, Lys 81 | E3 ligase Mule | Further polyubiquitylation by CHIP and protein degradation | [52] |
n.d. = not determined.
DOMINANT-NEGATIVE INTERFERENCE OF POLβ FUNCTION AND THE IMPACT OF TUMOR MUTATIONS
An essential aspect of any enzyme inhibitor to be used in cells or in vivo, as we have discussed throughout, is specificity and selectivity. Unfortunately, none of the small molecule inhibitors of Polβ discussed so far have been shown to be highly specific or selective. Many of these small molecule inhibitors can inhibit other DNA polymerases with equal or greater affinity, have multiple biological effects unrelated to Polβ or are directly cytotoxic, an effect not expected to result from Polβ inhibition since Polβ null cells are viable [13, 27, 108, 138].
Interestingly, Polβ expression or primary sequence appears to be altered in a significant percentage of human tumors so far evaluated [139]. High levels of Polβ expression have been demonstrated in several human cancers and tumor cell lines [140]. We, and others, have observed elevated expression of Polβ in gastrointestinal tumors and cancers from the esophagus, colon and pancreas [9, 141–143]. In addition, chronic myelogenous leukemia (CML) [144] and infection by human papillomavirus 16 (HPV16) [145] or Epstein-Barr virus (EBV) [146] leads to elevated expression of Polβ. In many cases, mutations within the Polβ coding region results in over-expression of dysfunctional Polβ proteins [8]. Overall, approximately 30% of human cancers express mutant or aberrant forms of Polβ proteins [8, 147–149], leading to genomic instability and possibly conferring a mutator phenotype to cells [108, 140, 150]. An updated list of disease-associated Polβ mutants is shown in Table 4. These range from single-point mutants such as the Leu22Pro mutant found in gastric cancer that is deficient in 5’dRP lyase activity to deletion mutants that function as dominant negative inhibitors of Polβ activity (Table 4). It is intriguing to consider the possibility that small molecules may be developed to specifically target cancer specific mutants of Polβ, although the likelihood of such inhibitors may be low. However, as we, and others, have shown, Polβ targeting by siRNA or shRNA gives rise to an almost complete loss of Polβ expression and increased sensitivity to DNA damaging agents [60, 62, 106, 151]. Hence, RNA interference could be utilized to specifically target mutant forms of Polβ, similar to that reported recently to target mutant forms of the Huntingtin alleles without affecting the normal allele [152]. Alternatively, it has been suggested that inhibiting the normal form of Polβ may be synthetically lethal with some cancer genotypes such as MSH2 mutations found in Hereditary NonPolyposis Colorectal Cancer (HNPCC) [153]. However, synthetic lethality of Polβ with a null mutation in MSH2 remains to be validated in a separate study.
Table 4.
Disease-Associated Polβ Mutants
| Mutation | Effect of mutation | Associated disease | Citation |
|---|---|---|---|
| Glu295Lys | Decreased polymerase activity, acts as a dominant negative | Gastric cancer | [210] |
| Leu22Pro | Loss of 5’dRP lyase activity | Gastric cancer | [211, 212] |
| Cys239Arg | Reduction in polymerase accuracy | Gastric cancer | [211] |
| Tyr39Cys | n.d. | ||
| Asn294Asp | n.d. | ||
| Asp160Asn | n.d. | ||
| Lys289Met | Reduction in polymerase accuracy | Colon cancer | [213, 214] |
| Δ208–236 | Decreased polymerase activity, acts as a dominant negative | Colon cancer | [8, 214] |
| Δ213–219 | n.d. | ||
| Δ249–262 | n.d. | ||
| Δ17 bp (frameshift and truncation = 209 amino acid protein) | n.d. | ||
| ΔT at codon 152 (frameshift and truncation = 162 amino acid protein) | n.d. | Prostate cancer | [8, 215] |
| Ile260Met | Misalignment-mediated errors in dipyrimidine sequences | Prostate cancer | [216] |
| Δ208–236 | Decreased polymerase activity, acts as a dominant negative | Lung cancer | [155, 214] |
| Δ546 bp (deletion of codons 20–202) | n.d. | Lung cancer | [155, 214] |
| Δ208–236 | Decreased polymerase activity, acts as a dominant negative | Breast cancer | [217] |
| Pro242Arg | n.d. | Breast cancer | [218] |
| Lys289Met | n.d. | ||
| Val215Pro | n.d. | Bladder cancer | [219] |
| Lys248Gln | n.d. | ||
| Ser229Pro | n.d. | ||
| A insertion at nucleotide 744 | n.d. | ||
| Arg183Gly | n.d. | Esophageal cancer | [220] |
| Glu177Stop | n.d. | ||
| Δ177–234 | n.d. | ||
| Gly179Arg | n.d. | ||
| Phe114Ser | n.d. | ||
| Gly118Glu | n.d. | ||
| Ile88Val | n.d. | ||
| Phe114Ser | n.d. | ||
| Lys167Ile | n.d. | ||
| Glu186Gly | n.d. | Esophageal cancer | [221] |
| Δ177–234 bp (frameshift) | n.d. | Esophageal cancer | [220, 221] |
| Δ208–236 | Decreased polymerase activity, acts as a dominant negative | Werner syndrome | [222] |
| Arg182Gly | n.d. | Cervical Cancer | [223] |
n.d. = not determined.
One interesting method that might be more amenable to a gene-therapy type approach is the identification and utilization of mutants of Polβ that function as dominantnegative inhibitors. Such an approach could take advantage of cell penetrating peptides for delivery of a highly specific Polβ and BER inhibitor [154]. The first report of a naturally occurring dominant-negative mutant of Polβ was from Banerjee and colleagues in which they identified a splicevariant of Polβ, missing 87 bp (exon 11) corresponding to amino acid residues 208–236 [155]. This mutant form of Polβ (PolβΔ87) is defective in BER [156] and functions as a dominant-negative inhibitor of Polβ [157]. Although suggested to be a cancer-specific form of Polβ [155], the PolβΔ87 mutant is expressed in both normal and tumor tissue [158].
Earlier, Wilson and colleagues demonstrated that the 14kDa N-terminal fragment of Polβ, corresponding to amino acid residues 1–140 (PolβN140), inhibited recombinant Polβ in vitro [159]. Interestingly, the inhibitory effect of PolβN140 was specific in that other N-terminal fragments, corresponding to fragment sizes of 8, 27 and 31 kDa did not inhibit the activity of Polβ and conversely, PolβN140 did not inhibit Polα [159]. Capitalizing on the dominant-negative phenotype of PolβN140, Vens and colleagues have shown repeatedly that this mutant form of Polβ can be utilized to specifically and selectively inhibit BER in human cells, likely by competing with the WT form of Polβ, as originally suggested by Wilson and colleagues [159]. Using this approach, they initially reported that PolβN140 revealed a role for Polβ in sensitization to ionizing radiation [61]. This dominant-negative mutant of Polβ acts as a radiosensitizer via XRCC1 dependent mechanisms that is independent of Polβ expression [160] and it’s action appears cell cycle dependent [161]. It is interesting to speculate that PolβN140 may function by interfering with XRCC1-dependent DNA repair complex formation. However, the mechanism of this interference is not obvious, as PolβN140 does not contain the XRCC1 binding domain. As might be expected, cell death due to radiation combined with expression of the dominantnegative PolβN140 correlates with accumulation of DNA double-strand breaks [161]. DNA double-strand break induction might result from interference with the repair of clustered lesions characteristic of ionizing radiation-induced damage [162]. We await the evaluation of the dominant-negative PolβN140 protein in xenograft studies and suggest that PolβN140 might be an excellent candidate for delivery using a cell penetrating peptide such as those described by Bitler and Schroeder [154].
INHIBITION OF POLβ-DEPENDENT 5’dRP LYASE ACTIVITY AND THE CELLULAR CONSEQUENCES
DNA Polβ has always been considered a “DNA Repair polymerase” [138] yet it was not until the development and characterization of Polβ knockout (KO) mouse embryonic fibroblasts (MEFs) that its role in BER was clearly defined where it was demonstrated that Polβ KO MEFs are sensitive to DNA alkylating agents due to a BER defect [13]. However, with the discovery that Polβ conducts two critical enzyme activities to complete BER, nucleotidyltransferase activity [18, 163] and 5’dRP lyase activity [20], it was important to determine the enzyme activity that was the most essential for BER and therefore if inhibited, could increase sensitivity to DNA alkylating agents. In a follow-up study, it was then determined that the alkylation sensitivity of Polβ KO MEFs was the result of a failure to repair the 5’dRP group, an intermediate in BER [27]. Subsequently, we have demonstrated that loss of Polβ-mediated 5’dRP lyase activity enhances sensitivity to the chemotherapeutic agent TMZ in human cells derived from breast and glioma tumors [60, 62, 106]. Although there appears to be several other enzymes that can repair the 5’dRP lesion [24, 29–33, 38, 39, 41, 47, 48], cellular hypersensitivity to alkylating agents can be achieved if one eliminates or attenuates the 5’dRP lyase activity of Polβ [27, 106]. In this final section, we present a summary of progress towards the development of effective inhibitors of the 5’dRP lyase activity of Polβ and discuss our recent insights into the cellular consequences that result from 5’dRP lyase inhibition.
Although not initially designed as such, many of the nucleotidyltransferase and DNA binding inhibitors discussed above also inhibit the 5’dRP lyase activity of Polβ. Earlier studies by Hecht, Sakaguchi and others (see above) focused on the polymerase activity of Polβ but the impact of many of these compounds on the associated 5’dRP lyase activity was not reported. However, it is likely that MA [73] and the sulfolipids (described above) may inhibit the 5’dRP lyase activity of Polβ since these and related compounds bind to the 8kDa domain of Polβ that encodes the 5’dRP lyase active site [82] (see Fig. (2)). Cloretazine is suggested to inhibit Polβ by modifying one of three cysteine amino acid residues. These all lie within the C-terminal or nucleotidyltrans-ferase domain and therefore if such modifications do occur, Cloretazine will likely effect the nucleotidyltransferase as opposed to the 5’dRP lyase activity of Polβ. On the other hand, LCA, originally identified as a DNA polymerase inhibitor, inhibits both the nucleotidyltransferase and the 5’dRP lyase activity of Polβ [107]. In addition, it was also determined that NSC666715 not only prevents binding to the damaged DNA but as a result, inhibits the 5’dRP lyase activity of Polβ [113]. From many of these studies, one may be able to extrapolate that tight binding to the 8kDa domain may be a signature or pre-requisite for inhibition of 5’dRP lyase activity.
In a completely surprising finding, Vijayanti and colleagues have reported that the dementia drugs donepezil hydrochloride, rivastigmine tartrate and Nootropil bind to the 8kDa domain of Polβ as determined in an in silico analysis to evaluate potential macromolecular docking sites on the protein [164]. Using in vitro DNA polymerase and 5’dRP lyase assays, they show no significant impact on polymerase activity but suggest that rivastigmine tartrate and Nootropil both have a significant impact on Polβ dependent 5’dRP lyase activity. However, there was little or no impact on a complete BER reaction containing DNA ligase. Additional studies are therefore suggested to validate the inhibitory effect of donepezil hydrochloride, rivastigmine tartrate and Nootropil on Polβ.
In what appears to be the first dedicated effort to identify and develop Polβ specific 5’dRP lyase inhibitors, Wilson and Hecht and colleagues have discovered a number of compounds that inhibit both the nucleotidyltransferase and 5’dRP lyase activity of Polβ, such as koetjapic acid (KJA), isolated from Sandoricum koetjape [70]. Using a bioassayguided fractionization approach and a standardized 5’dRP lyase activity assay, Hecht and Kingston identified a large number of 5’dRP lyase inhibitory compounds from natural products, including lupanetriterpinoids, (−)epicatechin, sesquiterpinoids, biscoumarin derivatives, plant sterols, oleananetriterpinoids, ursanetriterpenes and neolignans [165– 172]. In some cases, these compounds were reported to enhance or potentiate the cytotoxicity of the alkylating agent methyl methanesulfonate (MMS) [170], as might be expected from an inhibitor of the 5’dRP lyase activity of Polβ [27, 106]. The sulfolipid KN-208, a reported polymerase activity inhibitor (see above), enhanced cellular sensitivity to MMS [83] and is likely to inhibit the 5’dRP lyase activity of Polβ since the general class of sulfolipids are known to bind to the 8kDa domain of Polβ, although it has yet to be evaluated.
Selectivity for many of these compounds remains an issue since potentiation of MMS in a WT cell could result from the inhibition of many cellular functions. Wilson and Hecht approached this problem using a combined NMR, biochemical and cellular approach to identify lead compounds for specific inhibition of the 5’dRP lyase activity of Polβ [173]. In this initial study, they used their previously reported NMR structure of the 8kDa domain of Polβ [130, 131] to map the binding site and critical contacts (amino acid residues and chemical groups) for binding of the 5’dRP lyase inhibitor KJA [70]. Subsequently, thirty-four structurally similar compounds were analyzed using this NMR chemical shift mapping approach and ten compounds, including KJA, were identified that yielded appropriate (micromolar) binding constants. These and other 5’dRP lyase inhibitor compounds are listed in Table 5. To address the specificity issue, each of these compounds were then evaluated for the ability to potentiate MMS-induced cell death, comparing WT and Polβ KO MEFs [173], with the expectation that a highly specific Polβ 5’dRP lyase inhibitor would potentiate MMS in WT cells but would yield little or no potentiation in the Polβ KO cells. Any potentiation in the Polβ KO cells would likely be the result of non-specific or off-target effects. By comparing potentiation in both cell lines (WT and Polβ KO), Wilson and colleagues were then able to evaluate the specificity and efficacy of each compound by calculating an enhancement ratio (ER) [173], the most effective and specific compounds yielding the highest ER. By this criteria, it was determined that Pamoic Acid (PA) is the most effective and specific of the inhibitors tested, with a binding constant of 9 µM [173]. In line with this result, PA was also shown to inhibit both the nucleotidyltransferase and 5’dRP lyase activities of Polβ, when tested in an in vitro assay using purified, recombinant Polβ [173]. The binding interface of PA and the 8kDa domain of Polβ was then independently confirmed by Milon and colleagues [174] to further development of PA analogs and related compounds with greater affinity and selectivity for Polβ. Finally, a summary of the development and characterization of these compounds (Table 5) by Wilson and colleagues has also been reported [175].
Table 5.
DNA Polβ 5’dRP lyase inhibitors
| Compound Name | Abbreviation | Citation |
|---|---|---|
| Lithocholic acid | LCA | [107] |
| 4-chloro-N-[5-(4-chloroanilino)-1H-1,2,4-triazol-3-yl]-5-methyl-2-sulfanylbenzenesulfonamide | NSC666715 | [113] |
| Koetjapic acid | KJA* | [70, 173] |
| Biquinoline-dicarboxylic acid | BQD* | [173] |
| Naphthochrome green | NCG* | [173] |
| Mordant blue | MB* | [173] |
| Glycyrrhizic acid | GA* | [173] |
| 4,4′-(hexafluoroisopropylidene) bis(benzoic acid) | HFPB* | [173] |
| 3-(4-carboxyphenyl) 2,3-dihydrotrimethyl 1-indene-5-carboxylic acid | CPIC* | [173] |
| Carbenoxolone | CBX* | [173] |
| 4′4′-biphenyl dicarboxylic acid | BPDC* | [173] |
| Pamoic Acid | PA* | [173, 174] |
The structures have been reported – see [173].
The increased alkylating agent sensitivity of Polβ KO or deficient cells (or Polβ 5’dRP lyase deficient/inhibited cells) has been well documented [13, 27, 60, 62, 106, 108, 176, 177]. However, the mechanism of cell death initiated by alkylating agents in a Polβ null cell was only recently revealed. Using a series of human tumor cell lines depleted of Polβ (via RNA interference) together with isogenic lines complemented with WT and mutant forms of Polβ, we reported that failure to repair the 5’dRP lesion (termed BER Failure) initiates hyperactivation of PARP1/PARP2 leading to the depletion of NAD+/ATP pools, the release of the RAGE ligand HMGB1 and the onset of necrosis [106]. In fact, we find that cell death from Polβ inhibition (in concert with DNA damage) is highly dependent on the availability of cellular bioenergetic metabolites (NAD+ and NAD+ precursors) and the capacity of the NAD+ biosynthesis machinery [106]. In that vein, we suggest that the efficacy of Polβ inhibitors, especially 5’dRP lyase inhibitors, to potentiate DNA damage-induced cell death (e.g., anti-tumor effect) can be improved by simultaneous disruption or inhibition of cellular NAD+ biosynthesis [178].
SUMMARY
In summary, we have discussed past, present and future options for inhibiting Polβ as a means to enhance response to chemotherapy and radiation. The obvious target, to inhibit the nucleotidyltransferase activity of Polβ, has not resulted in any highly effective or specific inhibitors to date. The most effective compounds appear to be those that result in inhibition of both the nucleotidyltransferase and 5’dRP lyase activities of Polβ such as LCA, NSC666715 and PA. However, reported binding constants are still too high but NSC666715 and PA appear to provide a high level of specificity and offer promise, each targeting a unique site on Polβ. Future studies may also include novel targets such as the Polβ/XRCC1 interface. It has not yet been determined if interrupting the Polβ/XRCC1 interaction results in BER Failure or BER inhibition in cells, but studies are underway to determine if targeting BER-dependent protein-protein interfaces is a viable approach to inhibit BER. Another option is to modulate PTMs to inhibit Polβ. However, more information is needed about the functional impact of Polβ specific PTMs before considering this as a target. In addition, one report has proposed a potential for synthetic lethality between Polβ loss and mutations in MSH2, suggesting that targeting Polβ alone (in the absence of DNA damage) may be considered for MSH2-deficient tumors. Finally, as we discussed above, combinations of inhibitors may prove most effective in the short term. Since inhibiting Polβ triggers DNA damage-induced necrosis via NAD+ depletion [106], one might consider using a DNA damaging agent, a Polβ inhibitor and an NAD+ biosynthesis inhibitor. Such an approach might be most effective in tumors with defects in NAPRT1-mediated NAD+ biosynthesis to allow normal tissue rescue with nicotinic acid [179, 180].
ACKNOWLEDGEMENTS
We would like to extend special thanks to Lucas Santana dos Santos, University of Pittsburgh, for help with the PyMOL application. This work was supported by grants from the National Institutes of Health (NIH) [GM087798; CA148629] to RWS. Support was also provided by the University of Pittsburgh Department of Pharmacology & Chemical Biology to EMG and DS and a John S. Lazo Cancer Pharmacology Fellowship to EMG. Further, this publication was made possible by RI-INBRE Grant # P20RR016457 from the National Center for Research Resources (NCRR, NIH) awarded to KHA. It’s contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
ABBREVIATIONS
- 5’dRP
5’-deoxyribose phosphate
- APC
Adenomatous polyposis coli
- APE1
Apurinic/apyrimidinic endonuclease
- ATP
Adenosine-5’-triphosphate
- BER
Base excision repair
- BSA
Bovine serum albumen
- CML
Chronic myelogenous leukemia
- dNTP
Deoxynucleotide triphosphate
- dsDNA
Double-stranded DNA
- EBV
Epstein-Barr virus
- ER
Enhancement ratio
- HIV
Human Immunodeficiency virus
- IUPAC
International Union of Pure and Applied Chemistry
- KA-A
Kohamaic acid A
- KJA
Koetjapic acid
- KO
Knockout
- LCA
Lithocholic acid
- LigIIIa
DNA ligase IIIa
- MA
Masticadienonic acid
- MEF
Mouse embryonic fibroblast
- MGMT
O6-methylguanine-DNA methyltransferase
- MMS
Methyl methanesulfonate
- MNNG
N-methyl-N’-nitro-N-nitrosoguanidine
- MPG
Methylpurine (alkyladenine) DNA glycosylase
- NaBH4
Sodium borohydride
- NAD+
Nicotinamide adenine dinucleotide
- NMR
Nuclear magnetic resonance
- PA
Pamoic Acid
- PAR
Poly(ADP-ribose)
- PARG
Poly(ADP-ribose)glycohydrolase
- PARP1
Poly(ADP-ribose)polymerase-1
- PARP2
Poly(ADP-ribose)polymerase-2
- Pol
Polymerase
- Polβ
DNA polymerase β
- PolβΔ87
Polβ missing 87 bp (residues 208–236)
- PolβN140
Amino acid residues 1–140 of Polβ
- PTM
Post-translational modification
- SQAG
Sulfo-quinovosyl-acyl-glycerol
- SQMG
Sulfo-quinovosyl-monoacyl-glycerol
- ssDNA
Single-stranded DNA
- TMZ
Temozolomide
- uPAR
Urokinase-type plasminogen activator receptor
- WT
Wild type
REFERENCES
- 1.Burgers PM, Koonin EV, Bruford E, Blanco L, Burtis KC, Christman MF, Copeland WC, Friedberg EC, Hanaoka F, Hinkle DC, Lawrence CW, Nakanishi M, Ohmori H, Prakash L, Prakash S, Reynaud CA, Sugino A, Todo T, Wang Z, Weill JC, Woodgate R. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 2001;276(47):43487–43490. doi: 10.1074/jbc.R100056200. [DOI] [PubMed] [Google Scholar]
- 2.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv. Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 3.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair. 2007;6(6):695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127(4):709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 5.Fotiadou P, Henegariu O, Sweasy JB. DNA polymerase β interacts with TRF2 and induces telomere dysfunction in a murine mammary cell line. Cancer Res. 2004;64(11):3830–3837. doi: 10.1158/0008-5472.CAN-04-0136. [DOI] [PubMed] [Google Scholar]
- 6.Muftuoglu M, Wong HK, Imam SZ, Wilson DM, 3rd, Bohr VA, Opresko PL. Telomere repeat binding factor 2 interacts with base excision repair proteins and stimulates DNA synthesis by DNA polymerase beta. Cancer Res. 2006;66(1):113–124. doi: 10.1158/0008-5472.CAN-05-2742. [DOI] [PubMed] [Google Scholar]
- 7.Kidane D, Jonason AS, Gorton TS, Mihaylov I, Pan J, Keeney S, de Rooij DG, Ashley T, Keh A, Liu Y, Banerjee U, Zelterman D, Sweasy JB. DNA polymerase beta is critical for mouse meiotic synapsis. EMBO J. 2010;29(2):410–423. doi: 10.1038/emboj.2009.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starcevic D, Dalal S, Sweasy JB. Is there a link between DNA polymerase beta and cancer? Cell Cycle. 2004;3(8):998–1001. [PubMed] [Google Scholar]
- 9.Yoshizawa K, Jelezcova E, Brown AR, Foley JF, Nyska A, Cui X, Hofseth LJ, Maronpot RM, Wilson SH, Sepulveda AR, Sobol RW. Gastrointestinal Hyperplasia with Altered Expression of DNA Polymerase β. PLoS ONE. 2009;4(8):e6493. doi: 10.1371/journal.pone.0006493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu G, Herzig M, Rotrekl V, Walter CA. Base excision repair, aging and health span. Mech. Ageing Dev. 2008;129(7–8):366–382. doi: 10.1016/j.mad.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Copani A, Caraci F, Hoozemans JJ, Calafiore M, Sortino MA, Nicoletti F. The nature of the cell cycle in neurons: focus on a “non-canonical” pathway of DNA replication causally related to death. Biochim. Biophys. Acta. 2007;1772(4):409–412. doi: 10.1016/j.bbadis.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Copani A, Hoozemans JJ, Caraci F, Calafiore M, Van Haastert ES, Veerhuis R, Rozemuller AJ, Aronica E, Sortino MA, Nicoletti F. DNA polymerase-beta is expressed early in neurons of Alzheimer’s disease brain and is loaded into DNA replication forks in neurons challenged with beta-amyloid. J. Neurosci. 2006;26(43):10949–10957. doi: 10.1523/JNEUROSCI.2793-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-β in base-excision repair. Nature. 1996;379(6561):183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 14.Sobol RW, Temozolomide . In: Encyclopedia of Cancer. 2nd ed. M Schwab., editor. Berlin, Heidelberg, New York: Springer; 2009. [Google Scholar]
- 15.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291(5507):1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 16.Wilson SH, Sobol RW, Beard WA, Horton JK, Prasad R, Vande Berg BJ. DNA Polymerase β and Mammalian Base Excision Repair. Cold Spring Harb. Symp. Quant. Biol. 2001;65:143–155. doi: 10.1101/sqb.2000.65.143. [DOI] [PubMed] [Google Scholar]
- 17.Dantzer F, Ame JC, Schreiber V, Nakamura J, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006;409:493–510. doi: 10.1016/S0076-6879(05)09029-4. [DOI] [PubMed] [Google Scholar]
- 18.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase Beta. Chem. Rev. 2006;106(2):361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 19.Moon AF, Garcia-Diaz M, Batra VK, Beard WA, Bebenek K, Kunkel TA, Wilson SH, Pedersen LC. The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair (Amst) 2007;6(12):1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase β during DNA repair. Science. 1995;269(5224):699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto Y, Kim K, Katz DS, Feng JA. Catalytic center of DNA polymerase β for excision of deoxyribose phosphate groups. Biochemistry. 1998;37(18):6456–6464. doi: 10.1021/bi9727545. [DOI] [PubMed] [Google Scholar]
- 22.Prasad R, Batra VK, Yang XP, Krahn JM, Pedersen LC, Beard WA, Wilson SH. Structural insight into the DNA polymerase beta deoxyribose phosphate lyase mechanism. DNA Repair (Amst) 2005;4(12):1347–1357. doi: 10.1016/j.dnarep.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Prasad R, Beard WA, Chyan JY, Maciejewski MW, Mullen GP, Wilson SH. Functional analysis of the amino-terminal 8-kDa domain of DNA polymerase β as revealed by site-directed mutagenesis DNA binding and 5’-deoxyribose phosphate lyase activities. J. Biol. Chem. 1998;273(18):11121–11126. doi: 10.1074/jbc.273.18.11121. [DOI] [PubMed] [Google Scholar]
- 24.Prasad R, Beard WA, Strauss PR, Wilson SH. Human DNA polymerase β deoxyribose phosphate lyase Substrate specificity and catalytic mechanism. J. Biol. Chem. 1998;273(24):15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 25.Piersen CE, Prasad R, Wilson SH, Lloyd RS. Evidence for an imino intermediate in the DNA polymerase β deoxyribose phosphate excision reaction. J. Biol. Chem. 1996;271(30):17811–17815. doi: 10.1074/jbc.271.30.17811. [DOI] [PubMed] [Google Scholar]
- 26.Deterding LJ, Prasad R, Mullen GP, Wilson SH, Tomer KB. Mapping of the 5'-2-deoxyribose-5-phosphate lyase active site in DNA polymerase beta by mass spectrometry. J. Biol. Chem. 2000;275(14):10463–10471. doi: 10.1074/jbc.275.14.10463. [DOI] [PubMed] [Google Scholar]
- 27.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. The lyase activity of the DNA repair protein β-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405(6788):807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 28.Allinson SL, Dianova, Dianov GL. DNA polymerase β is the major dRP lyase involved in repair of oxidative base lesions in DNA by mammalian cell extracts. EMBO J. 2001;20(23):6919–6926. doi: 10.1093/emboj/20.23.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Diaz M, Bebenek K, Kunkel TA, Blanco L. Identification of an intrinsic 5'-deoxyribose-5-phosphate lyase activity in human DNA polymerase lambda: a possible role in base excision repair. J. Biol. Chem. 2001;276(37):34659–34663. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- 30.Bebenek K, Tissier A, Frank EG, McDonald JP, Prasad R, Wilson SH, Woodgate R, Kunkel TA. 5’-Deoxyribose phosphate lyase activity of human DNA polymerase iota in vitro . Science. 2001;291(5511):2156–2159. doi: 10.1126/science.1058386. [DOI] [PubMed] [Google Scholar]
- 31.Prasad R, Bebenek K, Hou E, Shock DD, Beard WA, Woodgate R, Kunkel TA, Wilson SH. Localization of the deoxyribose phosphate lyase active site in human DNA polymerase iota by controlled proteolysis. J. Biol. Chem. 2003;278(32):29649–29654. doi: 10.1074/jbc.M305399200. [DOI] [PubMed] [Google Scholar]
- 32.Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH. Human DNA polymerase theta possesses 5'-dRP lyase activity and functions in single-nucleotide base excision repair in vitro . Nucleic Acids Res. 2009;37(6):1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5'-deoxyribose phosphate lyase activity in human DNA polymerase γ and its role in mitochondrial base excision repair in vitro . Proc. Natl. Acad. Sci. USA. 1998;95(21):12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bogani F, Boehmer PE. The replicative DNA polymerase of herpes simplex virus 1 exhibits apurinic/apyrimidinic and 5’-deoxyribose phosphate lyase activities. Proc. Natl. Acad. Sci. USA. 2008;105(33):11709–11714. doi: 10.1073/pnas.0806375105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bogani F, Chua CN, Boehmer PE. Reconstitution of uracil DNA glycosylase-initiated base excision repair in herpes simplex virus-1. J. Biol. Chem. 2009;284(25):16784–16790. doi: 10.1074/jbc.M109.010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bogani F, Corredeira I, Fernandez V, Sattler U, Rutvisuttinunt W, Defais M, Boehmer PE. Association between the herpes simplex virus-1 DNA polymerase and uracil DNA glycosylase. J. Biol. Chem. 2010;285(36):27664–27672. doi: 10.1074/jbc.M110.131235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gellon L, Carson DR, Carson JP, Demple B. Intrinsic 5’-deoxyribose-5-phosphate lyase activity in Saccharomyces cerevisiae Trf4 protein with a possible role in base excision DNA repair. DNA Repair (Amst) 2008;7(2):187–198. doi: 10.1016/j.dnarep.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bandaru V, Zhao X, Newton MR, Burrows CJ, Wallace SS. Human endonuclease VIII-like (NEIL) proteins in the giant DNA Mimivirus. DNA Repair (Amst) 2007;6(11):1629–1641. doi: 10.1016/j.dnarep.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grin IR, Khodyreva SN, Nevinsky GA, Zharkov DO. Deoxyribophosphate lyase activity of mammalian endonuclease VIII-like proteins. FEBS Lett. 2006;580(20):4916–4922. doi: 10.1016/j.febslet.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 40.Alonso A, Terrados G, Picher AJ, Giraldo R, Blanco L, Larraga V. An intrinsic 5'-deoxyribose-5-phosphate lyase activity in DNA polymerase beta from Leishmania infantum supports a role in DNA repair. DNA Repair (Amst) 2006;5(1):89–101. doi: 10.1016/j.dnarep.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 41.Roberts SA, Strande N, Burkhalter MD, Strom C, Havener JM, Hasty P, Ramsden DA. Ku is a 5’-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464(7292):1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Escudero R, Garcia-Diaz M, Salas ML, Blanco L, Salas J. DNA polymerase X of African swine fever virus: insertion fidelity on gapped DNA substrates and AP lyase activity support a role in base excision repair of viral DNA. J. Mol. Biol. 2003;326(5):1403–1412. doi: 10.1016/s0022-2836(03)00019-6. [DOI] [PubMed] [Google Scholar]
- 43.Khairnar NP, Misra HS. DNA polymerase X from Deinococcus radiodurans implicated in bacterial tolerance to DNA damage is characterized as a short patch base excision repair polymerase. Microbiology. 2009;155(Pt. 9):3005–3014. doi: 10.1099/mic.0.029223-0. [DOI] [PubMed] [Google Scholar]
- 44.Saxowsky TT, Matsumoto Y, Englund PT. The mitochondrial DNA polymerase beta from Crithidia fasciculata has 5'-deoxyribose phosphate (dRP) lyase activity but is deficient in the release of dRP. J. Biol. Chem. 2002;277(40):37201–37206. doi: 10.1074/jbc.M206654200. [DOI] [PubMed] [Google Scholar]
- 45.Dianov G, Sedgwick B, Daly G, Olsson M, Lovett S, Lindahl T. Release of 5’-terminal deoxyribose-phosphate residues from incised abasic sites in DNA by the Escherichia coli RecJ protein. Nucleic Acids Res. 1994;22(6):993–998. doi: 10.1093/nar/22.6.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Graves RJ, Felzenszwalb I, Laval J, O’Connor TR. Excision of 5’-terminal deoxyribose phosphate from damaged DNA is catalyzed by the Fpg protein of Escherichia coli. J. Biol. Chem. 1992;267(20):14429–14435. [PubMed] [Google Scholar]
- 47.Summer H, Li O, Bao Q, Zhan L, Peter S, Sathiyanathan P, Henderson D, Klonisch T, Goodman SD, Droge P. HMGA2 exhibits dRP/AP site cleavage activity and protects cancer cells from DNA-damage-induced cytotoxicity during chemotherapy. Nucleic Acids Res. 2009;37(13):4371–4384. doi: 10.1093/nar/gkp375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prasad R, Liu Y, Deterding LJ, Poltoratsky VP, Kedar PS, Horton JK, Kanno S, Asagoshi K, Hou EW, Khodyreva SN, Lavrik OI, Tomer KB, Yasui A, Wilson SH. HMGB1 is a cofactor in mammalian base excision repair. Mol. Cell. 2007;27(5):829–841. doi: 10.1016/j.molcel.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Almeida KH, Sobol RW. Increased Specificity and Efficiency of Base Excision Repair through Complex Formation. In: Siede W, Doetsch PW, Kow YW, editors. DNA Damage Recognition. New York: Marcel Dekker Inc; 2005. pp. 33–64. [Google Scholar]
- 50.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA. 2004;101(33):12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hasan S, El-Andaloussi N, Hardeland U, Hassa PO, Burki C, Imhof R, Schar P, Hottiger MO. Acetylation regulates the DNA end-trimming activity of DNA polymerase β. Mol. Cell. 2002;10(5):1213–1222. doi: 10.1016/s1097-2765(02)00745-1. [DOI] [PubMed] [Google Scholar]
- 52.Parsons JL, Tait PS, Finch D, Dianova II, Edelmann MJ, Khoronenkova SV, Kessler BM, Sharma RA, McKenna WG, Dianov GL. Ubiquitin ligase ARF-BP1/Mule modulates base excision repair. EMBO J. 2009;28(20):3207–3215. doi: 10.1038/emboj.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sobol RW. CHIPping Away at Base Excision Repair. Mol. Cell. 2008;29(4):413–415. doi: 10.1016/j.molcel.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Parsons JL, Tait PS, Finch D, Dianova II, Allinson SL, Dianov GL. CHIP-Mediated Degradation and DNA Damage-Dependent Stabilization Regulate Base Excision Repair Proteins. Mol. Cell. 2008;29(4):477–487. doi: 10.1016/j.molcel.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 55.Berdis AJ. DNA polymerases as therapeutic targets. Biochemistry. 2008;47(32):8253–8260. doi: 10.1021/bi801179f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maga G, Hubscher U. Repair and translesion DNA polymerases as anticancer drug targets. Anticancer agents Med. Chem. 2008;8(4):431–447. doi: 10.2174/187152008784220348. [DOI] [PubMed] [Google Scholar]
- 57.Batista LF, Kaina B, Meneghini R, Menck CF. How DNA lesions are turned into powerful killing structures: insights from UV-induced apoptosis. Mutat Res. 2009;681(2–3):197–208. doi: 10.1016/j.mrrev.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 58.Huennekens FM. The methotrexate story: a paradigm for development of cancer chemotherapeutic agents. Adv. Enzyme Regul. 1994;34:397–419. doi: 10.1016/0065-2571(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 59.Sweasy JB, Lauper JM, Eckert KA. DNA polymerases and human diseases. Radiat. Res. 2006;166(5):693–714. doi: 10.1667/RR0706.1. [DOI] [PubMed] [Google Scholar]
- 60.Trivedi RN, Almeida KH, Fornsaglio JL, Schamus S, Sobol RW. The Role of Base Excision Repair in the Sensitivity and Resistance to Temozolomide Mediated Cell Death. Cancer Res. 2005;65(14):6394–6400. doi: 10.1158/0008-5472.CAN-05-0715. [DOI] [PubMed] [Google Scholar]
- 61.Vens C, Dahmen-Mooren E, Verwijs-Janssen M, Blyweert W, Graversen L, Bartelink H, Begg AC. The role of DNA polymerase beta in determining sensitivity to ionizing radiation in human tumor cells. Nucleic Acids Res. 2002;30(13):2995–3004. doi: 10.1093/nar/gkf403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trivedi RN, Wang XH, Jelezcova E, Goellner EM, Tang J, Sobol RW. Human methyl purine DNA glycosylase and DNA polymerase β expression collectively predict sensitivity to temozolomide. Mol. Pharmacol. 2008;74(2):505–516. doi: 10.1124/mol.108.045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma J, Starck SR, Hecht SM. DNA polymerase beta inhibitors from Tetracera boiviniana. J. Nat. Prod. 1999;62(12):1660–1663. doi: 10.1021/np990326p. [DOI] [PubMed] [Google Scholar]
- 64.Deng JZ, Starck SR, Hecht SM. DNA polymerase beta inhibitors from Baeckea gunniana. J. Nat. Prod. 1999;62(12):1624–1626. doi: 10.1021/np990240w. [DOI] [PubMed] [Google Scholar]
- 65.Deng JZ, Starck SR, Hecht SM. bis-5-Alkylresorcinols from Panopsis rubescens that inhibit DNA polymerase beta. J. Nat. Prod. 1999;62(3):477–480. doi: 10.1021/np980522g. [DOI] [PubMed] [Google Scholar]
- 66.Deng JZ, Starck SR, Hecht SM. Pentacyclic triterpenoids from Freziera sp that inhibit DNA polymerase beta. Bioorg. Med. Chem. 2000;8(1):247–250. doi: 10.1016/s0968-0896(99)00276-x. [DOI] [PubMed] [Google Scholar]
- 67.Deng JZ, Starck SR, Hecht SM, Ijames CF, Hemling ME. Harbinatic acid, a novel and potent DNA polymerase beta inhibitor from Hardwickia binata. J. Nat. Prod. 1999;62(7):1000–1002. doi: 10.1021/np990099r. [DOI] [PubMed] [Google Scholar]
- 68.Deng JZ, Starck SR, Sun DA, Sabat M, Hecht SM. A new 7,8-euphadien-type triterpenoid from Brackenridgea nitida and Bleasdalea bleasdalei that inhibits DNA polymerase beta. J. Nat. Prod. 2000;63(10):1356–1360. doi: 10.1021/np000129m. [DOI] [PubMed] [Google Scholar]
- 69.Starck SR, Deng JZ, Hecht SM. Naturally occurring alkylresorcinols that mediate DNA damage and inhibit its repair. Biochemistry. 2000;39(9):2413–2419. doi: 10.1021/bi991509d. [DOI] [PubMed] [Google Scholar]
- 70.Sun DA, Starck SR, Locke EP, Hecht SM. DNA polymerase beta inhibitors from Sandoricum koetjape. J. Nat. Prod. 1999;62(8):1110–1113. doi: 10.1021/np990104r. [DOI] [PubMed] [Google Scholar]
- 71.Deng JZ, Starck SR, Li S, Hecht SM. (+)-Myristinins A and D from Knema elegans, which inhibit DNA polymerase beta and cleave DNA. J. Nat. Prod. 2005;68(11):1625–1628. doi: 10.1021/np058064g. [DOI] [PubMed] [Google Scholar]
- 72.Maloney DJ, Deng JZ, Starck SR, Gao Z, Hecht SM. (+)-Myristinin A, a naturally occurring DNA polymerase beta inhibitor and potent DNA-damaging agent. J. Am. Chem. Soc. 2005;127(12):4140–4141. doi: 10.1021/ja042727j. [DOI] [PubMed] [Google Scholar]
- 73.Boudsocq F, Benaim P, Canitrot Y, Knibiehler M, Ausseil F, Capp JP, Bieth A, Long C, David B, Shevelev I, Frierich-Heinecken E, Hubscher U, Amalric F, Massiot G, Hoffmann JS, Cazaux C. Modulation of cellular response to cisplatin by a novel inhibitor of DNA polymerase beta. Mol. Pharmacol. 2005;67(5):1485–1492. doi: 10.1124/mol.104.001776. [DOI] [PubMed] [Google Scholar]
- 74.Nakamura R, Takeuchi R, Kuramochi K, Mizushina Y, Ishimaru C, Takakusagi Y, Takemura M, Kobayashi S, Yoshida H, Sugawara F, Sakaguchi K. Chemical properties of fatty acid derivatives as inhibitors of DNA polymerases. Org. Biomol. Chem. 2007;5(24):3912–3921. doi: 10.1039/b710944j. [DOI] [PubMed] [Google Scholar]
- 75.Kimura T, Nishida M, Kuramochi K, Sugawara F, Yoshida H, Mizushina Y. Novel azaphilones, kasanosins A and B, which are specific inhibitors of eukaryotic DNA polymerases beta and lambda from Talaromyces sp. Bioorg. Med. Chem. 2008;16(8):4594–4599. doi: 10.1016/j.bmc.2008.02.037. [DOI] [PubMed] [Google Scholar]
- 76.Naganuma M, Nishida M, Kuramochi K, Sugawara F, Yoshida H, Mizushina Y. 1-deoxyrubralactone, a novel specific inhibitor of families X and Y of eukaryotic DNA polymerases from a fungal strain derived from sea algae. Bioorg. Med. Chem. 2008;16(6):2939–2944. doi: 10.1016/j.bmc.2007.12.044. [DOI] [PubMed] [Google Scholar]
- 77.Mizushina Y, Watanabe I, Ohta K, Takemura M, Sahara H, Takahashi N, Gasa S, Sugawara F, Matsukage A, Yoshida S, Sakaguchi K. Studies on inhibitors of mammalian DNA polymerase alpha and beta: sulfolipids from a pteridophyte, Athyrium niponicum. Biochem. Pharmacol. 1998;55(4):537–541. doi: 10.1016/s0006-2952(97)00536-4. [DOI] [PubMed] [Google Scholar]
- 78.Hanashima S, Mizushina Y, Ohta K, Yamazaki T, Sugawara F, Sakaguchi K. Structure-activity relationship of a novel group of mammalian DNA polymerase inhibitors, synthetic sulfoquinovosylacylglycerols. Jpn. J. Cancer Res. 2000;91(10):1073–1083. doi: 10.1111/j.1349-7006.2000.tb00887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hanashima S, Mizushina Y, Yamazaki T, Ohta K, Takahashi S, Sahara H, Sakaguchi K, Sugawar F. Synthesis of sulfoquinovosylacylglycerols, inhibitors of eukaryotic DNA polymerase alpha and beta. Bioorg. Med. Chem. 2001;9(2):367–376. doi: 10.1016/s0968-0896(00)00252-2. [DOI] [PubMed] [Google Scholar]
- 80.Mizushina Y, Kasai N, Iijima H, Sugawara F, Yoshida H, Sakaguchi K. Sulfo-quinovosyl-acyl-glycerol (SQAG), a eukaryotic DNA polymerase inhibitor and anti-cancer agent. Curr. Med. Chem. Anticancer Agents. 2005;5(6):613–625. doi: 10.2174/156801105774574685. [DOI] [PubMed] [Google Scholar]
- 81.Ohta K, Mizushina Y, Hirata N, Takemura M, Sugawara F, Matsukage A, Yoshida S, Sakaguchi K. Action of a new mammalian DNA polymerase inhibitor, sulfoquinovosyldiacylglycerol. Biol. Pharm. Bull. 1999;22(2):111–116. doi: 10.1248/bpb.22.111. [DOI] [PubMed] [Google Scholar]
- 82.Kasai N, Mizushina Y, Murata H, Yamazaki T, Ohkubo T, Sakaguchi K, Sugawara F. Sulfoquinovosylmonoacylglycerol inhibitory mode analysis of rat DNA polymerase beta. FEBS J. 2005;272(17):4349–4361. doi: 10.1111/j.1742-4658.2005.04848.x. [DOI] [PubMed] [Google Scholar]
- 83.Ogawa A, Murate T, Izuta S, Takemura M, Furuta K, Kobayashi J, Kamikawa T, Nimura Y, Yoshida S. Sulfated glycoglycerolipid from archaebacterium inhibits eukaryotic DNA polymerase alpha, beta and retroviral reverse transcriptase and affects methyl methanesulfonate cytotoxicity. Int. J. Cancer. 1998;76(4):512–518. doi: 10.1002/(sici)1097-0215(19980518)76:4<512::aid-ijc12>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 84.Murakami C, Takemura M, Yoshida H, Sugawara F, Sakaguchi K, Mizushina Y. Analysis of cell cycle regulation by 1-mono-O-acyl-3-O-(alpha-D-sulfoquinovosyl)-glyceride (SQMG), an inhibitor of eukaryotic DNA polymerases. Biochem. Pharmacol. 2003;66(4):541–550. doi: 10.1016/s0006-2952(03)00345-9. [DOI] [PubMed] [Google Scholar]
- 85.Mori Y, Sahara H, Matsumoto K, Takahashi N, Yamazaki T, Ohta K, Aoki S, Miura M, Sugawara F, Sakaguchi K, Sato N. Downregulation of Tie2 gene by a novel antitumor sulfolipid, 3'-sulfoquinovosyl-1'-monoacylglycerol, targeting angiogenesis. Cancer Sci. 2008;99(5):1063–1070. doi: 10.1111/j.1349-7006.2008.00785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sakimoto I, Ohta K, Yamazaki T, Ohtani S, Sahara H, Sugawara F, Sakaguchi K, Miura M. Alphasulfoquinovosylmonoacylglycerol is a novel potent radiosensitizer targeting tumor angiogenesis. Cancer Res. 2006;66(4):2287–2295. doi: 10.1158/0008-5472.CAN-05-2209. [DOI] [PubMed] [Google Scholar]
- 87.Mizushina Y, Murakami C, Yogi K, Ueda K, Ishidoh T, Takemura M, Perpelescu M, Suzuki M, Oshige M, Yamaguchi T, Saneyoshi M, Yoshida H, Sakaguchi K. Kohamaic acid A, a novel sesterterpenic acid, inhibits activities of DNA polymerases from deuterostomes. Biochim. Biophys. Acta. 2003;1648(1–2):55–61. doi: 10.1016/s1570-9639(03)00108-0. [DOI] [PubMed] [Google Scholar]
- 88.Mizushina Y, Manita D, Takeuchi T, Sugawara F, Kumamoto-Yonezawa Y, Matsui Y, Takemura M, Sasaki M, Yoshida H, Takikawa H. The inhibitory action of kohamaic acid A derivatives on mammalian DNA polymerase beta. Molecules. 2009;14(1):102–121. doi: 10.3390/molecules14010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takikawa H, Kamatani N, Nakanishi K, Tashiro T, Sasaki M, Yoshida H, Mizushina Y. Synthetic studies on kohamaic acids: synthesis of structurally simplified analogs of kohamaic acid A. Biosci. Biotechnol. Biochem. 2008;72(11):3071–3074. doi: 10.1271/bbb.80629. [DOI] [PubMed] [Google Scholar]
- 90.Mizushina Y, Ohkubo T, Date T, Yamaguchi T, Saneyoshi M, Sugawara F, Sakaguchi K. Mode analysis of a fatty acid molecule binding to the N-terminal 8-kDa domain of DNA polymerase beta A 1:1 complex and binding surface. J. Biol. Chem. 1999;274(36):25599–25607. doi: 10.1074/jbc.274.36.25599. [DOI] [PubMed] [Google Scholar]
- 91.Frederick AM, Davis ML, Rice KP. Inhibition of human DNA polymerase beta activity by the anticancer prodrug Cloretazine. Biochem. Biophys. Res. Commun. 2009;378(3):419–423. doi: 10.1016/j.bbrc.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krishna G, Hodnick WF, Lang W, Lin X, Karra S, Mao J, Almassian B. Pharmaceutical development and manufacturing of a parenteral formulation of a novel antitumor agent, VNP40101M. AAPS PharmSciTech. 2001;2(3):E14. doi: 10.1208/pt020314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Deisseroth A, Farrell A, Justice R, Kane R, Sridhara R, Chen H, He K, Pazdur R. Toxicity of laromustine plus highdose cytarabine in patients with relapsed acute myeloid leukemia. Blood. 2010;115(2):430. doi: 10.1182/blood-2009-09-244236. [DOI] [PubMed] [Google Scholar]
- 94.Giles F, Vey N, DeAngelo D, Seiter K, Stock W, Stuart R, Boskovic D, Pigneux A, Tallman M, Brandwein J, Kell J, Robak T, Staib P, Thomas X, Cahill A, Albitar M, O’Brien S. Phase 3 randomized, placebo-controlled, double-blind study of high-dose continuous infusion cytarabine alone or with laromustine (VNP40101M) in patients with acute myeloid leukemia in first relapse. Blood. 2009;114(19):4027–4033. doi: 10.1182/blood-2009-06-229351. [DOI] [PubMed] [Google Scholar]
- 95.Schiller GJ, O’Brien SM, Pigneux A, Deangelo DJ, Vey N, Kell J, Solomon S, Stuart RK, Karsten V, Cahill AL, Albitar MX, Giles FJ. Single-agent laromustine, a novel alkylating agent, has significant activity in older patients with previously untreated poor-risk acute myeloid leukemia. J. Clin. Oncol. 2010;28(5):815–821. doi: 10.1200/JCO.2009.24.2008. [DOI] [PubMed] [Google Scholar]
- 96.Ishiguro K, Shyam K, Penketh PG, Sartorelli AC. Role of O6-alkylguanine-DNA alkyltransferase in the cytotoxic activity of cloretazine. Mol. Cancer Ther. 2005;4(11):1755–1763. doi: 10.1158/1535-7163.MCT-05-0169. [DOI] [PubMed] [Google Scholar]
- 97.Rice KP, Penketh PG, Shyam K, Sartorelli AC. Differential inhibition of cellular glutathione reductase activity by isocyanates generated from the antitumor prodrugs Cloretazine and BCNU. Biochem. Pharmacol. 2005;69(10):1463–1472. doi: 10.1016/j.bcp.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 98.Baba M, Schols D, Nakashima H, Pauwels R, Parmentier G, Meijer DK, De Ckercq E. Selective activity of several cholic acid derivatives against human immunodeficiency virus replication in vitro . J. Acquir. Immune Defic. Syndr. 1989;2(3):264–271. [PubMed] [Google Scholar]
- 99.Ogawa A, Murate T, Suzuki M, Nimura Y, Yoshida S. Lithocholic acid, a putative tumor promoter, inhibits mammalian DNA polymerase beta. Jpn. J. Cancer Res. 1998;89(11):1154–1159. doi: 10.1111/j.1349-7006.1998.tb00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pool-Zobel BL, Leucht U. Induction of DNA damage by risk factors of colon cancer in human colon cells derived from biopsies. Mutat. Res. 1997;375(2):105–115. doi: 10.1016/s0027-5107(97)00006-7. [DOI] [PubMed] [Google Scholar]
- 101.Zusman I, Zimber A. Ultrastructural changes in rat colorectal epithelium and tumors after treatment with N-methyl-N'-nitro-N-nitrosoguanidine and secondary bile acids. Acta Anat. (Basel) 1991;141(3):282–288. doi: 10.1159/000147135. [DOI] [PubMed] [Google Scholar]
- 102.Mizushina Y, Kasai N, Miura K, Hanashima S, Takemura M, Yoshida H, Sugawara F, Sakaguchi K. Structural relationship of lithocholic acid derivatives binding to the N-terminal 8-kDa domain of DNA polymerase beta. Biochemistry. 2004;43(33):10669–10677. doi: 10.1021/bi049307r. [DOI] [PubMed] [Google Scholar]
- 103.Watanabe M, Hanashima S, Mizushina Y, Yoshida H, Oshige M, Sakaguchi K, Sugawara F. Biotinylated lithocholic acids for affinity chromatography of mammalian DNA polymerases alpha and beta. Bioorg. Med. Chem. Lett. 2002;12(3):287–290. doi: 10.1016/s0960-894x(01)00725-9. [DOI] [PubMed] [Google Scholar]
- 104.Kuramochi K, Haruyama T, Takeuchi R, Sunoki T, Watanabe M, Oshige M, Kobayashi S, Sakaguchi K, Sugawara F. Affinity capture of a mammalian DNA polymerase beta by inhibitors immobilized to resins used in solid-phase organic synthesis. Bioconjug. Chem. 2005;16(1):97–104. doi: 10.1021/bc0497970. [DOI] [PubMed] [Google Scholar]
- 105.Mizushina Y, Kasai N, Sugawara F, Iida A, Yoshida H, Sakaguchi K. Three-dimensional structural model analysis of the binding site of lithocholic acid, an inhibitor of DNA polymerase beta and DNA topoisomerase II. J. Biochem. 2001;130(5):657–664. doi: 10.1093/oxfordjournals.jbchem.a003031. [DOI] [PubMed] [Google Scholar]
- 106.Tang J, Goellner EM, Wang XW, Trivedi RN, st. Croix CM, Jelezcova E, Svilar D, Brown AR, Sobol RW. Bioenergetic Metabolites Regulate Base Excision Repair-Dependent Cell Death in Response to DNA Damage. Mol. Cancer Res. 2010;8(1):67–79. doi: 10.1158/1541-7786.MCR-09-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stachelek GC, Dalal S, Donigan KA, Campisi Hegan D, Sweasy JB, Glazer PM. Potentiation of temozolomide cytotoxicity by inhibition of DNA polymerase beta is accentuated by BRCA2 mutation. Cancer Res. 2010;70(1):409–417. doi: 10.1158/0008-5472.CAN-09-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sobol RW, Kartalou M, Almeida KH, Joyce DF, Engelward BP, Horton JK, Prasad R, Samson LD, Wilson SH. Base Excision Repair Intermediates Induce p53-independent Cytotoxic and Genotoxic Responses. J. Biol. Chem. 2003;278(41):39951–39959. doi: 10.1074/jbc.M306592200. [DOI] [PubMed] [Google Scholar]
- 109.Ruiz-Gaspa S, Guanabens N, Enjuanes A, Peris P, Martinez-Ferrer A, de Osaba MJ, Gonzalez B, Alvarez L, Monegal A, Combalia A, Pares A. Lithocholic acid downregulates vitamin D effects in human osteoblasts. Eur. J. Clin. Invest. 2010;40(1):25–34. doi: 10.1111/j.1365-2362.2009.02230.x. [DOI] [PubMed] [Google Scholar]
- 110.Sun J, Mustafi R, Cerda S, Chumsangsri A, Xia YR, Li YC, Bissonnette M. Lithocholic acid down-regulation of NF-kappaB activity through vitamin D receptor in colonic cancer cells. J. Steroid Biochem. Mol. Biol. 2008;111(1–2):37–40. doi: 10.1016/j.jsbmb.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Baek MK, Park JS, Park JH, Kim MH, Kim HD, Bae WK, Chung IJ, Shin BA, Jung YD. Lithocholic acid upregulates uPAR and cell invasiveness via MAPK and AP-1 signaling in colon cancer cells. Cancer Lett. 2010;290(1):123–128. doi: 10.1016/j.canlet.2009.08.030. [DOI] [PubMed] [Google Scholar]
- 112.Goldberg AA, Richard VR, Kyryakov P, Bourque SD, Beach A, Burstein MT, Glebov A, Koupaki O, Boukh-Viner T, Gregg C, Juneau M, English AM, Thomas DY, Titorenko VI. Chemical genetic screen identifies lithocholic acid as an anti-aging compound that extends yeast chronological life span in a TOR-independent manner, by modulating housekeeping longevity assurance processes. Aging (Albany NY) 2010;2(7):393–414. doi: 10.18632/aging.100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jaiswal AS, Banerjee S, Panda H, Bulkin CD, Izumi T, Sarkar FH, Ostrov DA, Narayan S. A novel inhibitor of DNA polymerase beta enhances the ability of temozolomide to impair the growth of colon cancer cells. Mol. Cancer Res. 2009;7(12):1973–1983. doi: 10.1158/1541-7786.MCR-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Brzozowski Z. 2-Mercapto-N-(azolyl)benzenesulphonamides. I. Synthesis of N-(1,1-dioxo-1,4,2-benzodithiazin-3-yl)guanidines and their transformations into 2-mercapto-N-(5-amino-1,2,4-triazol-3-yl) benzenesulphonamide derivatives with potential anti-HIV or anticancer activity. Acta Pol. Pharm. 1995;52(2):91–101. [PubMed] [Google Scholar]
- 115.Brzozowski Z. 2-Mercapto-N-(azolyl)benzenesulfonamides. V. Syntheses, anti-HIV and anticancer activity of some 4-chloro-2-mercapto-5-methyl-N-(1,2,4- triazolo[4,3-a]pyrid-3-yl)benzenesulfonamides. Acta Pol. Pharm. 1998;55(5):375–379. [PubMed] [Google Scholar]
- 116.Neamati N, Mazumder A, Zhao H, Sunder S, Burke TR, Jr., Schultz RJ, Pommier Y. Diarylsulfones, a novel class of human immunodeficiency virus type 1 integrase inhibitors. Antimicrob. Agents Chemother. 1997;41(2):385–393. doi: 10.1128/aac.41.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jaiswal AS, Narayan S. A novel function of adenomatous polyposis coli (APC) in regulating DNA repair. Cancer Lett. 2008;271(2):272–280. doi: 10.1016/j.canlet.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Narayan S, Jaiswal AS, Balusu R. Tumor suppressor APC blocks DNA polymerase beta-dependent strand displacement synthesis during long patch but not short patch base excision repair and increases sensitivity to methylmethane sulfonate. J. Biol. Chem. 2005;280(8):6942–6949. doi: 10.1074/jbc.M409200200. [DOI] [PubMed] [Google Scholar]
- 119.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: Implications for DNA damage recognition, removal, and repair. DNA Repair. 2007;6(4):410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 120.Chen JY, Mamidipalli S, Huan T. HAPPI: an online database of comprehensive human annotated and predicted protein interactions. BMC Genomics. 2009;10(Suppl. 1):S16. doi: 10.1186/1471-2164-10-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463(7280):457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Reddy CD, Dasgupta P, Saikumar P, Dudek H, Rauscher FJ, 3rd, Reddy EP. Mutational analysis of Max: role of basic, helix-loop-helix/leucine zipper domains in DNA binding, dimerization and regulation of Myc-mediated transcriptional activation. Oncogene. 1992;7(10):2085–2092. [PubMed] [Google Scholar]
- 123.Berg T. Small-Molecule Modulators of c-Myc/Max and Max/Max Interactions. Curr. Top. Microbiol. Immunol. 2010;348:139–149. doi: 10.1007/82_2010_90. [DOI] [PubMed] [Google Scholar]
- 124.Clausen DM, Guo J, Parise RA, Beumer JH, Egorin MJ, Lazo JS, Prochownik EV, Eiseman JL. In vitro cytotoxicity and in vivo efficacy, pharmacokinetics, and metabolism of 10074-G5, a novel small-molecule inhibitor of c-Myc/Max dimerization. J. Pharmacol. Exp. Ther. 2010;335(3):715–727. doi: 10.1124/jpet.110.170555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Flygare JA, Fairbrother WJ. Small-molecule pan-IAP antagonists: a patent review. Expert Opin. Ther. Pat. 2010;20(2):251–267. doi: 10.1517/13543770903567077. [DOI] [PubMed] [Google Scholar]
- 126.Chen DJ, Huerta S. Smac mimetics as new cancer therapeutics. Anticancer drugs. 2009;20(8):646–658. doi: 10.1097/CAD.0b013e32832ced78. [DOI] [PubMed] [Google Scholar]
- 127.Cragg MS, Harris C, Strasser A, Scott CL. Unleashing the power of inhibitors of oncogenic kinases through BH3 mimetics. Nat. Rev. Cancer. 2009;9(5):321–326. doi: 10.1038/nrc2615. [DOI] [PubMed] [Google Scholar]
- 128.Lessene G, Czabotar PE, Colman PM. BCL-2 family antagonists for cancer therapy. Nat. Rev. Drug Discov. 2008;7(12):989–1000. doi: 10.1038/nrd2658. [DOI] [PubMed] [Google Scholar]
- 129.Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase β and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro . Nucleic Acids Res. 1996;24(22):4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Marintchev A, Robertson A, Dimitriadis EK, Prasad R, Wilson SH, Mullen GP. Domain specific interaction in the XRCC1-DNA polymerase β complex. Nucleic Acids Res. 2000;28(10):2049–2059. doi: 10.1093/nar/28.10.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gryk MR, Marintchev A, Maciejewski MW, Robertson A, Wilson SH, Mullen GP. Mapping of the interaction interface of DNA polymerase β with XRCC1. Structure (Camb) 2002;10(12):1709–1720. doi: 10.1016/s0969-2126(02)00908-5. [DOI] [PubMed] [Google Scholar]
- 132.Cuneo MJ, London RE. Oxidation state of the XRCC1 N-terminal domain regulates DNA polymerase beta binding affinity. Proc. Natl. Acad. Sci. USA. 2010;107(15):6805–6810. doi: 10.1073/pnas.0914077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase β and the XRCC1 protein. EMBO J. 1996;15(23):6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 134.Marintchev A, Gryk MR, Mullen GP. Site-directed mutagenesis analysis of the structural interaction of the singlestrand-break repair protein, X-ray cross-complementing group 1, with DNA polymerase β. Nucleic Acids Res. 2003;31(2):580–588. doi: 10.1093/nar/gkg159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dianova II, Sleeth KM, Allinson SL, Parsons JL, Breslin C, Caldecott KW, Dianov GL. XRCC1-DNA polymerase beta interaction is required for efficient base excision repair. Nucleic Acids Res. 2004;32(8):2550–2555. doi: 10.1093/nar/gkh567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Parsons JL, Dianova II, Allinson SL, Dianov GL. DNA polymerase beta promotes recruitment of DNA ligase III alpha-XRCC1 to sites of base excision repair. Biochemistry. 2005;44(31):10613–10619. doi: 10.1021/bi050085m. [DOI] [PubMed] [Google Scholar]
- 137.Wong HK, Wilson DM., 3rd XRCC1 and DNA polymerase beta interaction contributes to cellular alkylating-agent resistance and single-strand break repair. J. Cell Biochem. 2005;95(4):794–804. doi: 10.1002/jcb.20448. [DOI] [PubMed] [Google Scholar]
- 138.Sobol RW. DNA polymerase β null mouse embryonic fibroblasts harbor a homozygous null mutation in DNA polymerase iota. DNA Repair (Amst) 2007;6(1):3–7. doi: 10.1016/j.dnarep.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Albertella MR, Lau A, O'Connor MJ. The overexpression of specialized DNA polymerases in cancer. DNA Repair (Amst) 2005;4(5):583–593. doi: 10.1016/j.dnarep.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 140.Srivastava DK, Husain I, Arteaga CL, Wilson SH. DNA polymerase β expression differences in selected human tumors and cell lines. Carcinogenesis. 1999;20(6):1049–1054. doi: 10.1093/carcin/20.6.1049. [DOI] [PubMed] [Google Scholar]
- 141.Dong ZM, Zheng NG, Wu JL, Li SK, Wang YL. Difference in expression level and localization of DNA polymerase beta among human esophageal cancer focus, adjacent and corresponding normal tissues. Dis. Esophagus. 2006;19(3):172–176. doi: 10.1111/j.1442-2050.2006.00560.x. [DOI] [PubMed] [Google Scholar]
- 142.Yu J, Mallon MA, Zhang W, Freimuth RR, Marsh S, Watson MA, Goodfellow PJ, McLeod HL. DNA repair pathway profiling and microsatellite instability in colorectal cancer. Clin. Cancer Res. 2006;12(17):5104–5111. doi: 10.1158/1078-0432.CCR-06-0547. [DOI] [PubMed] [Google Scholar]
- 143.Fan R, Kumaravel TS, Jalali F, Marrano P, Squire JA, Bristow RG. Defective DNA strand break repair after DNA damage in prostate cancer cells: implications for genetic instability and prostate cancer progression. Cancer Res. 2004;64(23):8526–8533. doi: 10.1158/0008-5472.CAN-04-1601. [DOI] [PubMed] [Google Scholar]
- 144.Canitrot Y, Laurent G, Astarie-Dequeker C, Bordier C, Cazaux C, Hoffmann JS. Enhanced expression and activity of DNA polymerase beta in chronic myelogenous leukemia. Anticancer Res. 2006;26(1B):523–525. [PubMed] [Google Scholar]
- 145.Liu SN, Bai WY, Frye RM, 2nd, Hou L, Zhang B. Specific up-regulation of DNA polymerase by human papillomavirus 16. Chin. Med. Sci. J. 2008;23(2):108–112. [PubMed] [Google Scholar]
- 146.Faumont N, Le Clorennec NC, Teira P, Goormachtigh G, Coll J, Canitrot Y, Cazaux C, Hoffmann JS, Brousset P, Delsol G, Feuillard J, Meggetto F. Regulation of DNA polymerase beta by the LMP1 oncoprotein of EBV through the nuclear factor-kappaB pathway. Cancer Res. 2009;69(12):5177–5185. doi: 10.1158/0008-5472.CAN-08-2866. [DOI] [PubMed] [Google Scholar]
- 147.Michiels S, Laplanche A, Boulet T, Dessen P, Guillonneau B, Mejean A, Desgrandchamps F, Lathrop M, Sarasin A, Benhamou S. Genetic polymorphisms in 85 DNA repair genes and bladder cancer risk. Carcinogenesis. 2009;30(5):763–768. doi: 10.1093/carcin/bgp046. [DOI] [PubMed] [Google Scholar]
- 148.Li D, Suzuki H, Liu B, Morris J, Liu J, Okazaki T, Li Y, Chang P, Abbruzzese JL. DNA repair gene polymorphisms and risk of pancreatic cancer. Clin. Cancer Res. 2009;15(2):740–746. doi: 10.1158/1078-0432.CCR-08-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li D, Li Y, Jiao L, Chang DZ, Beinart G, Wolff RA, Evans DB, Hassan MM, Abbruzzese JL. Effects of base excision repair gene polymorphisms on pancreatic cancer survival. Int. J. Cancer. 2007;120(8):1748–1754. doi: 10.1002/ijc.22301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sobol RW, Watson DE, Nakamura J, Yakes FM, Hou E, Horton JK, Ladapo J, Van Houten B, Swenberg JA, Tindall KR, Samson LD, Wilson SH. Mutations associated with base excision repair deficiency and methylation-induced genotoxic stress. Proc. Natl. Acad. Sci. USA. 2002;99(10):6860–6865. doi: 10.1073/pnas.092662499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Polosina YY, Rosenquist TA, Grollman AP, Miller H. ‘Knock down’ of DNA polymerase β by RNA interference: recapitulation of null phenotype. DNA Repair (Amst) 2004;3(11):1469–1474. doi: 10.1016/j.dnarep.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 152.Pfister EL, Kennington L, Straubhaar J, Wagh S, Liu W, DiFiglia M, Landwehrmeyer B, Vonsattel JP, Zamore PD, Aronin N. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr. Biol. 2009;19(9):774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Martin SA, McCabe N, Mullarkey M, Cummins R, Burgess DJ, Nakabeppu Y, Oka S, Kay E, Lord CJ, Ashworth A. DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH2 or MLH1. Cancer Cell. 2010;17(3):235–248. doi: 10.1016/j.ccr.2009.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bitler BG, Schroeder JA. Anti-cancer therapies that utilize cell penetrating peptides. Recent Pat. Anticancer Drug Discov. 2010;5(2):99–108. doi: 10.2174/157489210790936252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bhattacharyya N, Chen HC, Comhair S, Erzurum SC, Banerjee S. Variant forms of DNA polymerase β in primary lung carcinomas. DNA Cell Biology. 1999;18(7):549–554. doi: 10.1089/104454999315097. [DOI] [PubMed] [Google Scholar]
- 156.Bhattacharyya N, Banerjee T, Patel U, Banerjee S. Impaired repair activity of a truncated DNA polymerase β protein. Life Sciences. 2001;69(3):271–280. doi: 10.1016/s0024-3205(01)01120-1. [DOI] [PubMed] [Google Scholar]
- 157.Bhattacharyya N, Banerjee S. A variant of DNA polymerase β acts as a dominant negative mutant. Proc. Natl. Acad. Sci. USA. 1997;94(19):10324–10329. doi: 10.1073/pnas.94.19.10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bu D, Cler LR, Lewis CM, Euhus DM. A variant of DNA polymerase beta is not cancer specific. J. Invest. Surg. 2004;17(6):327–331. doi: 10.1080/08941930490524372. [DOI] [PubMed] [Google Scholar]
- 159.Husain I, Morton BS, Beard WA, Singhal RK, Prasad R, Wilson SH, Besterman JM. Specific inhibition of DNA polymerase beta by its 14 kDa domain: role of single- and doublestranded DNA binding and 5'-phosphate recognition. Nucleic Acids Res. 1995;23(9):1597–1603. doi: 10.1093/nar/23.9.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Neijenhuis S, Begg AC, Vens C. Radiosensitization by a dominant negative to DNA polymerase beta is DNA polymerase beta-independent and XRCC1-dependent. Radiother Oncol. 2005;76(2):123–128. doi: 10.1016/j.radonc.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 161.Neijenhuis S, Verwijs-Janssen M, Kasten-Pisula U, Rumping G, Borgmann K, Dikomey E, Begg AC, Vens C. Mechanism of cell killing after ionizing radiation by a dominant negative DNA polymerase beta. DNA Repair (Amst) 2009;8(3):336–346. doi: 10.1016/j.dnarep.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 162.Vens C, Begg AC. Targeting base excision repair as a sensitization strategy in radiotherapy. Semin. Radiat. Oncol. 2010;20(4):241–249. doi: 10.1016/j.semradonc.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 163.Beard WA, Wilson SH. Structural design of a eukaryotic DNA repair polymerase: DNA polymerase beta. Mutat. Res. 2000;460(3–4):231–244. doi: 10.1016/s0921-8777(00)00029-x. [DOI] [PubMed] [Google Scholar]
- 164.Vyjayanti VN, Chary NS, Rao KS. On the inhibitory affect of some dementia drugs on DNA polymerase Beta activity. Neurochem. Res. 2008;33(11):2187–2196. doi: 10.1007/s11064-007-9587-3. [DOI] [PubMed] [Google Scholar]
- 165.Li SS, Gao Z, Feng X, Jones SH, Hecht SM. Plant sterols as selective DNA polymerase beta lyase inhibitors and potentiators of bleomycin cytotoxicity. Bioorg. Med. Chem. 2004;12(15):4253–4258. doi: 10.1016/j.bmc.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 166.Prakash Chaturvedula VS, Gao Z, Hecht SM, Jones SH, Kingston DG. A new acylated oleanane triterpenoid from Couepia polyandra that inhibits the lyase activity of DNA polymerase β. J. Nat. Prod. 2003;66(11):1463–1465. doi: 10.1021/np0301893. [DOI] [PubMed] [Google Scholar]
- 167.Cao S, Gao Z, Thomas SJ, Hecht SM, Lazo JS, Kingston DG. Marine sesquiterpenoids that inhibit the lyase activity of DNA polymerase beta. J. Nat. Prod. 2004;67(10):1716–1718. doi: 10.1021/np049849+. [DOI] [PubMed] [Google Scholar]
- 168.Chaturvedula VS, Gao Z, Jones SH, Feng X, Hecht SM, Kingston DG. A new ursane triterpene from Monochaetum vulcanicum that inhibits DNA polymerase beta lyase. J. Nat. Prod. 2004;67(5):899–901. doi: 10.1021/np030531b. [DOI] [PubMed] [Google Scholar]
- 169.Chaturvedula VS, Zhou BN, Gao Z, Thomas SJ, Hecht SM, Kingston DG. New lupane triterpenoids from Solidago canadensis that inhibit the lyase activity of DNA polymerase beta. Bioorg. Med. Chem. 2004;12(23):6271–6275. doi: 10.1016/j.bmc.2004.08.048. [DOI] [PubMed] [Google Scholar]
- 170.Feng X, Gao Z, Li S, Jones SH, Hecht SM. DNA polymerase beta lyase inhibitors from Maytenus putterlickoides. J. Nat. Prod. 2004;67(10):1744–1747. doi: 10.1021/np040057p. [DOI] [PubMed] [Google Scholar]
- 171.Li SS, Gao Z, Feng X, Hecht SM. Biscoumarin derivatives from Edgeworthia gardneri that inhibit the lyase activity of DNA polymerase beta. J. Nat. Prod. 2004;67(9):1608–1610. doi: 10.1021/np040127s. [DOI] [PubMed] [Google Scholar]
- 172.Prakash Chaturvedula VS, Hecht SM, Gao Z, Jones SH, Feng X, Kingston DG. New neolignans that inhibit DNA polymerase beta lyase. J. Nat. Prod. 2004;67(6):964–967. doi: 10.1021/np030507y. [DOI] [PubMed] [Google Scholar]
- 173.Hu HY, Horton JK, Gryk MR, Prasad R, Naron JM, Sun DA, Hecht SM, Wilson SH, Mullen GP. Identification of small molecule synthetic inhibitors of DNA polymerase beta by NMR chemical shift mapping. J. Biol. Chem. 2004;279(38):39736–39744. doi: 10.1074/jbc.M402842200. [DOI] [PubMed] [Google Scholar]
- 174.Hazan C, Boudsocq F, Gervais V, Saurel O, Ciais M, Cazaux C, Czaplicki J, Milon A. Structural insights on the pamoic acid and the 8 kDa domain of DNA polymerase beta complex: towards the design of higher-affinity inhibitors. BMC Struct. Biol. 2008;8:22. doi: 10.1186/1472-6807-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wilson SH, Beard WA, Shock DD, Batra VK, Cavanaugh NA, Prasad R, Hou EW, Liu Y, Asagoshi K, Horton JK, Stefanick DF, Kedar PS, Carrozza MJ, Masaoka A, Heacock ML. Base excision repair and design of small molecule inhibitors of human DNA polymerase beta. Cell Mol. Life Sci. 2010;67(21):3633–3647. doi: 10.1007/s00018-010-0489-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Horton JK, Joyce-Gray DF, Pachkowski BF, Swenberg JA, Wilson SH. Hypersensitivity of DNA polymerase beta null mouse fibroblasts reflects accumulation of cytotoxic repair intermediates from site-specific alkyl DNA lesions. DNA Repair (Amst) 2003;2(1):27–48. doi: 10.1016/s1568-7864(02)00184-2. [DOI] [PubMed] [Google Scholar]
- 177.Horton JK, Prasad R, Hou E, Wilson SH. Protection against methylation-induced cytotoxicity by DNA polymerase β-dependent long patch base excision repair. J. Biol. Chem. 2000;275(3):2211–2218. doi: 10.1074/jbc.275.3.2211. [DOI] [PubMed] [Google Scholar]
- 178.Goellner EM, Grimme B, Brown AR, Lin YC, Wang XH, Sugrue KF, Mitchell L, Trivedi RN, Tang JB, Sobol RW. Overcoming Temozolomide Resistance in Glioblastoma via Dual Inhibition of NAD+ Biosynthesis and Base Excision Repair. Cancer Res. 2011;71(6):2308–2317. doi: 10.1158/0008-5472.CAN-10-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Watson M, Roulston A, Belec L, Billot X, Marcellus R, Bedard D, Bernier C, Branchaud S, Chan H, Dairi K, Gilbert K, Goulet D, Gratton MO, Isakau H, Jang A, Khadir A, Koch E, Lavoie M, Lawless M, Nguyen M, Paquette D, Turcotte E, Berger A, Mitchell M, Shore GC, Beauparlant P. The small molecule GMX1778 is a potent inhibitor of NAD+ biosynthesis: strategy for enhanced therapy in nicotinic acid phosphoribosyltransferase 1-deficient tumors. Mol. Cell Biol. 2009;29(21):5872–5888. doi: 10.1128/MCB.00112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Olesen UH, Thougaard AV, Jensen PB, Sehested M. A preclinical study on the rescue of normal tissue by nicotinic acid in high-dose treatment with APO866, a specific nicotinamide phosphoribosyltransferase inhibitor. Mol. Cancer Ther. 2010;9(6):1609–1617. doi: 10.1158/1535-7163.MCT-09-1130. [DOI] [PubMed] [Google Scholar]
- 181.Cannizzaro LA, Bollum FJ, Huebner K, Croce CM, Cheung LC, Xu X, Hecht BK, Hecht F, Chang LM. Chromosome sublocalization of a cDNA for human DNA polymerase-beta to 8p11----p12. Cytogenet. Cell Genet. 1988;47(3):121–124. doi: 10.1159/000132527. [DOI] [PubMed] [Google Scholar]
- 182.McBride OW, Zmudzka BZ, Wilson SH. Chromosomal location of the human gene for DNA polymerase beta. Proc. Natl. Acad. Sci. USA. 1987;84(2):503–507. doi: 10.1073/pnas.84.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chyan YJ, Ackerman S, Shepherd NS, McBride OW, Widen SG, Wilson SH, Wood TG. The human DNA polymerase beta gene structure. Evidence of alternative splicing in gene expression. Nucleic Acids Res. 1994;22(14):2719–2725. doi: 10.1093/nar/22.14.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Braithwaite EK, Kedar PS, Stumpo DJ, Bertocci B, Freedman JH, Samson LD, Wilson SH. DNA polymerases beta and lambda mediate overlapping and independent roles in base excision repair in mouse embryonic fibroblasts. PLoS ONE. 2010;5(8):e12229. doi: 10.1371/journal.pone.0012229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Balusu R, Jaiswal AS, Armas ML, Kundu CN, Bloom LB, Narayan S. Structure/function analysis of the interaction of adenomatous polyposis coli with DNA polymerase beta and its implications for base excision repair. Biochemistry. 2007;46(49):13961–13974. doi: 10.1021/bi701632e. [DOI] [PubMed] [Google Scholar]
- 186.Jaiswal AS, Balusu R, Armas ML, Kundu CN, Narayan S. Mechanism of adenomatous polyposis coli (APC)-mediated blockage of long-patch base excision repair. Biochemistry. 2006;45(51):15903–15914. doi: 10.1021/bi0607958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bennett RAO, Wilson DM, 3rd, Wong D, Demple B. Interaction of human apurinic endonuclease and DNA polymerase β in the base excision repair pathway. Proc. Natl. Acad. Sci. USA. 1997;94(14):7166–7169. doi: 10.1073/pnas.94.14.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Balakrishnan L, Brandt PD, Lindsey-Boltz LA, Sancar A, Bambara RA. Long patch base excision repair proceeds via coordinated stimulation of the multienzyme DNA repair complex. J. Biol. Chem. 2009;284(22):15158–15172. doi: 10.1074/jbc.M109.000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Prasad R, Dianov GL, Bohr VA, Wilson SH. FEN1 stimulation of DNA polymerase β mediates an excision step in mammalian long patch base excision repair. J. Biol. Chem. 2000;275(6):4460–4466. doi: 10.1074/jbc.275.6.4460. [DOI] [PubMed] [Google Scholar]
- 190.Takahashi A, Yamakawa N, Mori E, Ohnishi K, Yokota S, Sugo N, Aratani Y, Koyama H, Ohnishi T. Development of thermotolerance requires interaction between polymerase-beta and heat shock proteins. Cancer Sci. 2008;99(5):973–978. doi: 10.1111/j.1349-7006.2008.00759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Mendez F, Kozin E, Bases R. Heat shock protein 70 stimulation of the deoxyribonucleic acid base excision repair enzyme polymerase beta. Cell stress chaperones. 2003;8(2):153–161. doi: 10.1379/1466-1268(2003)008<0153:hspsot>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Prasad R, Singhal RK, Srivastava DK, Molina JT, Tomkinson AE, Wilson SH. Specific interaction of DNA polymerase β and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J. Biol. Chem. 1996;271(27):16000–16007. doi: 10.1074/jbc.271.27.16000. [DOI] [PubMed] [Google Scholar]
- 193.Wang L, Bhattacharyya N, Chelsea DM, Escobar PF, Banerjee S. A novel nuclear protein, MGC5306 interacts with DNA polymerase beta and has a potential role in cellular phenotype. Cancer Res. 2004;64(21):7673–7677. doi: 10.1158/0008-5472.CAN-04-2801. [DOI] [PubMed] [Google Scholar]
- 194.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122(6):957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 195.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004;15(2):209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 196.Das A, Wiederhold L, Leppard JB, Kedar P, Prasad R, Wang H, Boldogh I, Karimi-Busheri F, Weinfeld M, Tomkinson AE, Wilson SH, Mitra S, Hazra TK. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: Evidence for a repair complex in human cells. DNA Repair (Amst) 2006;5(12):1439–1448. doi: 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhou J, Ahn J, Wilson SH, Prives C. A role for p53 in base excision repair. EMBO J. 2001;20(4):914–923. doi: 10.1093/emboj/20.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Prasad R, Lavrik OI, Kim SJ, Kedar P, Yang XP, Vande Berg BJ, Wilson SH. DNA polymerase β-mediated long patch base excision repair Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. J. Biol. Chem. 2001;276(35):32411–32414. doi: 10.1074/jbc.C100292200. [DOI] [PubMed] [Google Scholar]
- 199.Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002;277(25):23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 200.Kedar PS, Kim SJ, Robertson A, Hou E, Prasad R, Horton JK, Wilson SH. Direct interaction between mammalian DNA polymerase β and proliferating cell nuclear antigen. J. Biol. Chem. 2002;277(34):31115–31123. doi: 10.1074/jbc.M201497200. [DOI] [PubMed] [Google Scholar]
- 201.Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104(1):107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 202.El-Andaloussi N, Valovka T, Toueille M, Hassa PO, Gehrig P, Covic M, Hubscher U, Hottiger MO. Methylation of DNA polymerase beta by protein arginine methyltransferase 1 regulates its binding to proliferating cell nuclear antigen. FASEB J. 2007;21(1):26–34. doi: 10.1096/fj.06-6194com. [DOI] [PubMed] [Google Scholar]
- 203.El-Andaloussi N, Valovka T, Toueille M, Steinacher R, Focke F, Gehrig P, Covic M, Hassa PO, Schar P, Hubscher U, Hottiger MO. Arginine methylation regulates DNA polymerase β. Mol. Cell. 2006;22(1):51–62. doi: 10.1016/j.molcel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 204.Toueille M, El-Andaloussi N, Frouin I, Freire R, Funk D, Shevelev I, Friedrich-Heineken E, Villani G, Hottiger MO, Hubscher U. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase β and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 2004;32(11):3316–3324. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Maga G, Crespan E, Wimmer U, van Loon B, Amoroso A, Mondello C, Belgiovine C, Ferrari E, Locatelli G, Villani G, Hubscher U. Replication protein A and proliferating cell nuclear antigen coordinate DNA polymerase selection in 8-oxo-guanine repair. Proc. Natl. Acad. Sci. USA. 2008;105(52):20689–20694. doi: 10.1073/pnas.0811241106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Harrigan JA, Opresko PL, von Kobbe C, Kedar PS, Prasad R, Wilson SH, Bohr VA. The Werner syndrome protein stimulates DNA polymerase β strand displacement synthesis via its helicase activity. J. Biol. Chem. 2003;278(25):22686–22695. doi: 10.1074/jbc.M213103200. [DOI] [PubMed] [Google Scholar]
- 207.Tokui T, Inagaki M, Nishizawa K, Yatani R, Kusagawa M, Ajiro K, Nishimoto Y, Date T, Matsukage A. Inactivation of DNA polymerase beta by in vitro phosphorylation with protein kinase C. J. Biol. Chem. 1991;266(17):10820–10824. [PubMed] [Google Scholar]
- 208.Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, Possemato A, Nardone J, Innocenti G, Wetzel R, Wang Y, MacNeill J, Mitchell J, Gygi SP, Rush J, Polakiewicz RD, Comb MJ. Signaling networks assembled by oncogenic EGFR and c-Met. Proc. Natl. Acad. Sci. USA. 2008;105(2):692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Phosphosite. PhosphositePlus. cited; Available from: http://www.phosphosite.org.
- 210.Lang T, Dalal S, Chikova A, Dimaio D, Sweasy JB. The E295K DNA polymerase beta gastric cancer-associated variant interferes with base excision repair and induces cellular transformation. Mol. Cell Biol. 2007;27(15):5587–5596. doi: 10.1128/MCB.01883-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Iwanaga A, Ouchida M, Miyazaki K, Hori K, Mukai T. Functional mutation of DNA polymerase β found in human gastric cancer: inability of the base excision repair in vitro . Mutat. Res. 1999;435(2):121–128. doi: 10.1016/s0921-8777(99)00036-1. [DOI] [PubMed] [Google Scholar]
- 212.Dalal S, Chikova A, Jaeger J, Sweasy JB. The Leu22Pro tumor-associated variant of DNA polymerase beta is dRP lyase deficient. Nucleic Acids Res. 2008;36(2):411–422. doi: 10.1093/nar/gkm1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Lang T, Maitra M, Starcevic D, Li SX, Sweasy JB. A DNA polymerase beta mutant from colon cancer cells induces mutations. Proc. Natl. Acad. Sci. USA. 2004;101(16):6074–6079. doi: 10.1073/pnas.0308571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang L, Patel U, Ghosh L, Banerjee S. DNA polymerase β mutations in human colorectal cancer. Cancer Res. 1992;52(17):4824–4827. [PubMed] [Google Scholar]
- 215.Dobashi Y, Shuin T, Tsuruga H, Uemura H, Torigoe S, Kubota Y. DNA polymerase β gene mutation in human prostate cancer. Cancer Res. 1994;54(11):2827–2829. [PubMed] [Google Scholar]
- 216.Dalal S, Hile S, Eckert KA, Sun KW, Starcevic D, Sweasy JB. Prostate-cancer-associated I260M variant of DNA polymerase beta is a sequence-specific mutator. Biochemistry. 2005;44(48):15664–15673. doi: 10.1021/bi051179z. [DOI] [PubMed] [Google Scholar]
- 217.Bhattacharyya N, Chen HC, Grundfest-Broniatowski S, Banerjee S. Alteration of hMSH2 and DNA polymerase β genes in breast carcinomas and fibroadenomas. Biochem. Biophys. Res. Commun. 1999;259(2):429–435. doi: 10.1006/bbrc.1999.0791. [DOI] [PubMed] [Google Scholar]
- 218.Sliwinski T, Ziemba P, Morawiec Z, Kowalski M, Zadrozny M, Blasiak J. Polymorphisms of the DNA polymerase beta gene in breast cancer. Breast Cancer Res. Treat. 2007;103(2):161–166. doi: 10.1007/s10549-006-9357-y. [DOI] [PubMed] [Google Scholar]
- 219.Matsuzaki J, Dobashi Y, Miyamoto H, Ikeda I, Fujinami K, Shuin T, Kubota Y. DNA polymerase β gene mutations in human bladder cancer. Mol. Carcinog. 1996;15(1):38–43. doi: 10.1002/(SICI)1098-2744(199601)15:1<38::AID-MC6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 220.Dong Z, Zhao G, Zhao Q, Yang H, Xue L, Tan X, Zheng N. A study of DNA polymerase beta mutation in human esophageal cancer. Zhonghua Yi Xue Za Zhi. 2002;82(13):899–902. [PubMed] [Google Scholar]
- 221.Zhao GQ, Wang T, Zhao Q, Yang HY, Tan XH, Dong ZM. Mutation of DNA polymerase beta in esophageal carcinoma of different regions. World J. Gastroenterol. 2005;11(30):4618–4622. doi: 10.3748/wjg.v11.i30.4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sadakane Y, Maeda K, Kuroda Y, Hori K. Identification of mutations in DNA polymerase beta mRNAs from patients with Werner syndrome. Biochem. Biophys. Res. Commun. 1994;200(1):219–225. doi: 10.1006/bbrc.1994.1437. [DOI] [PubMed] [Google Scholar]
- 223.Han LP, Qiao YH, Dong ZM, Shi HR, Zhao GQ, Liu D. Study on DNA polymerase beta gene mutation in human cervical cancer. Zhonghua fu chan ke za zhi. 2003;38(10):618–620. [PubMed] [Google Scholar]