Figure 2. Number of model states.
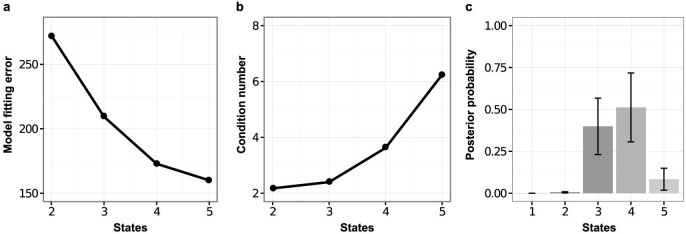
Models having n states were fitted to gene expression time-course data from a secondary MEF-based reprogramming system due to Samavarchi-Tehrani et al.18. (a), Model fitting error (quantified as the squared difference between output of the fitted model and the observed data) decreases with n but at the cost of introducing artifactual, non-distinct states as shown, (b), by an increase in the condition number that quantifies linear dependence between the state-specific gene expression signatures, i.e., mutual similarity of the n state-specific profiles. (c), A Bayesian model selection approach was used to score models with different numbers of states in terms of their posterior probability. The four-state model has the highest posterior probability, while there is negligible support for a 2-state, or single step, model. Taken together these results suggest that a four-state model strikes a good balance between fit-to-data and model complexity.
