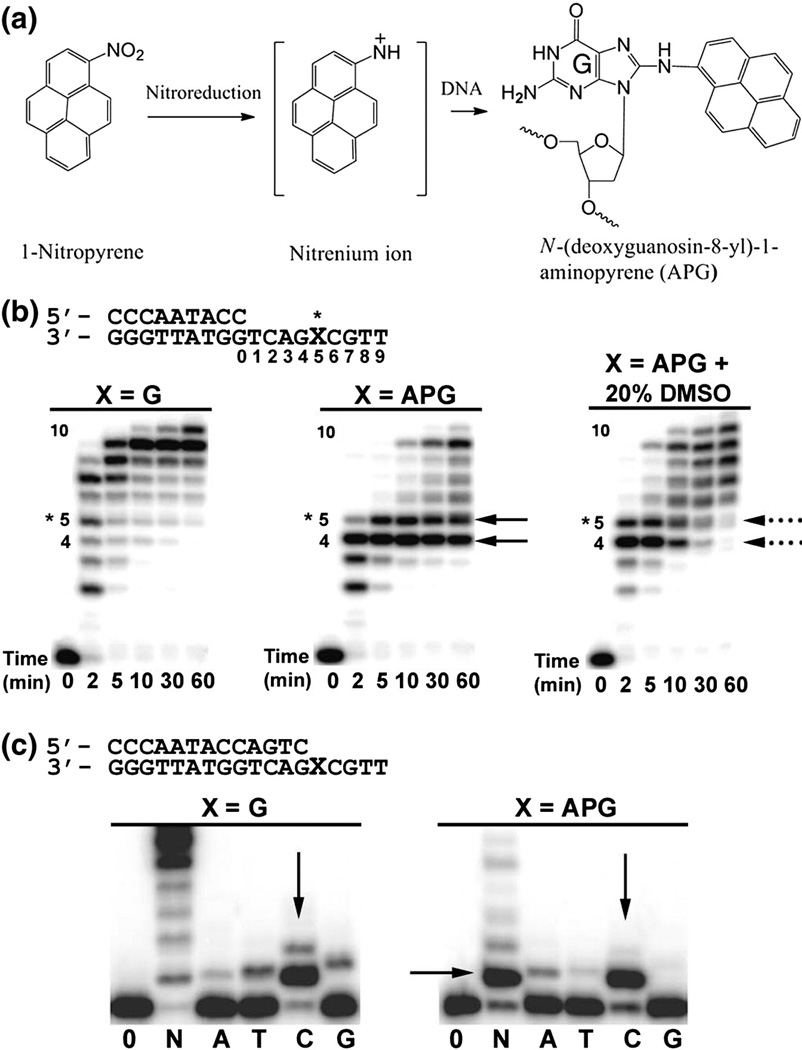
Fig. 1.
Formation of the APG adduct and Dpo4 bypass activity. (a) The 1-NP compound is reduced in the cell to generate one or more reactive species that react with the C8 position of guanine nucleotides, producing the APG lesion. (b) Running-start primer extension assays with Dpo4 and undamaged G or the APG lesion. Dpo4 was incubated with DNA substrates and reacted in the presence of all four nucleotides at various time points, indicated under each lane. Replication stalling at APG pre-insertion (position 4) and APG insertion (position 5) are denoted with black arrows. The resolving of stalling bands in the presence of 20% (v/v) DMSO is indicated with black broken lines. (c) Standing-start primer extension assays with Dpo4 and undamaged G or the APG lesion at the first replication position. Dpo4 was incubated with DNA substrates and reacted with all four nucleotides (N) or individual nucleotides (A, T, C, and G) for either 2 min for undamaged G or 30 min for the APG lesion. Vertical arrows indicate nucleotide preferences opposite G and the APG lesion. DNA substrates are shown above the gels.