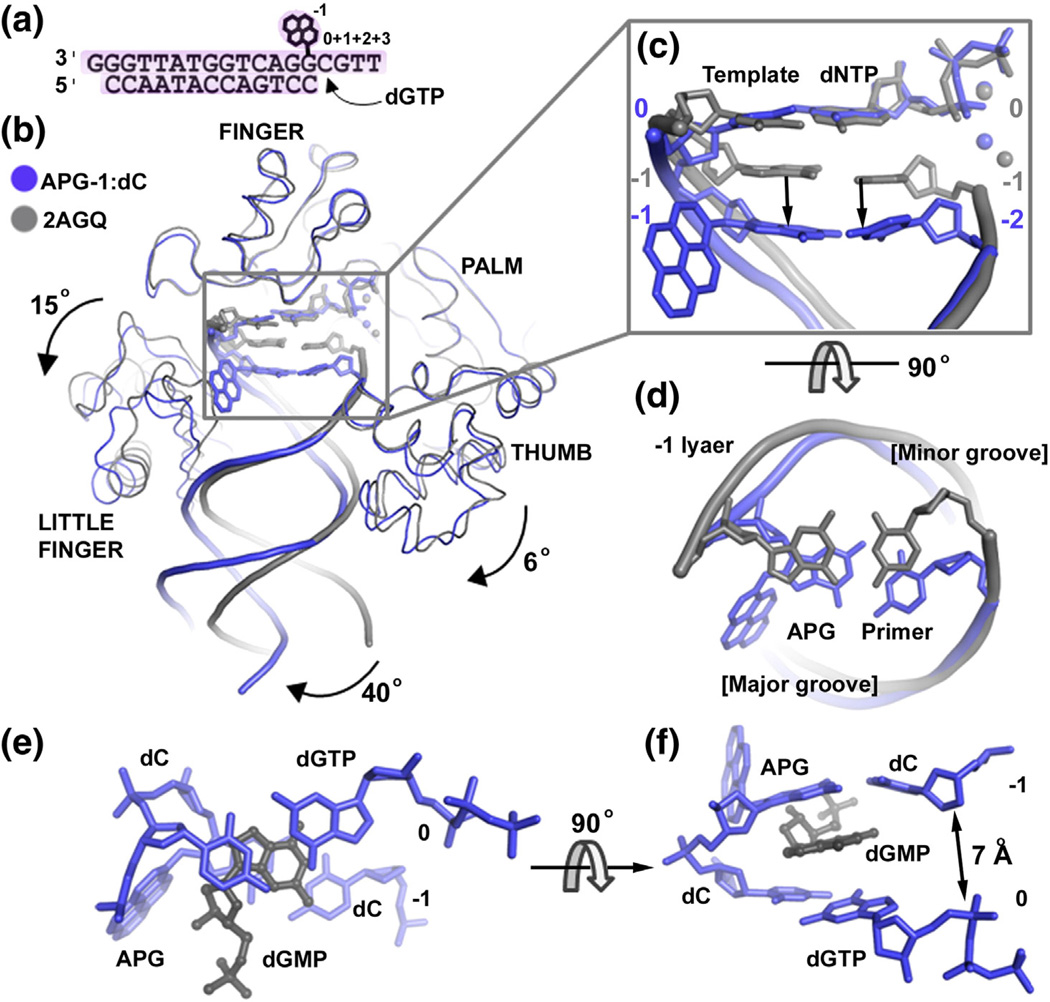
Fig. 4.
APG-1 in comparison to the undamaged DNA ternary structure. (a) DNA substrate used for post-insertion crystallization. (b) Superposition of the APG-1 structure (blue) with a previously solved Dpo4 ternary complex with undamaged DNA (PDB ID: 2AGQ; gray). The dGMP in APG-1 has been removed for clarity. Individual domains are labeled and arrows indicate movement relative to Dpo4 in complex with undamaged DNA. The APG lesion and incoming nucleotides are shown in stick model with coordinated metal ions as spheres. (c) Zoom in view of the superimposed active sites showing how the APG:dC base pair moves down into a non-standard position. Top view shows the major and minor groove sides of the DNA helix. (d) The −1 base pairs (upstream to the replicating base pair) in the APG-1 and undamaged DNA structures. (e and f) The 0 and −1 base pairs with dGMP in the APG-1 structure.