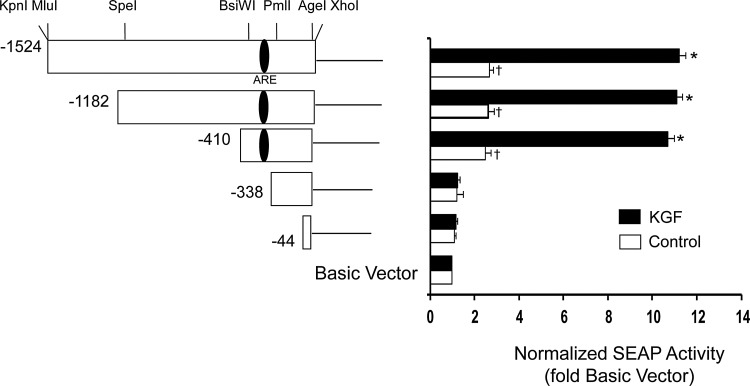
FIG. 2.
Basal and KGF-inducible activity of PRDX6 promoter deletions. Left panel, a series of constructs were generated by restriction enzyme digestion of the 1.5 kb DNA fragment derived from the 5′-flanking region of PRDX6 in the promoterless pSEAP2-Basic vector. Numbers marking the 5′ ends of the deletions are relative to the translational start. The approximate location of the putative antioxidant response element (ARE) is shown within the promoter. Right panel, A549 cells were transiently transfected with reporter vectors and then incubated for 24 h, followed by 20 ng/ml of KGF treatment (or no treatment in control samples). SEAP activity was determined 24 h later. Mean±SE (n=3) are shown. *p<0.01 compared with vehicle control, †p<0.05 compared with pSEAP2-basic vector (no insert).