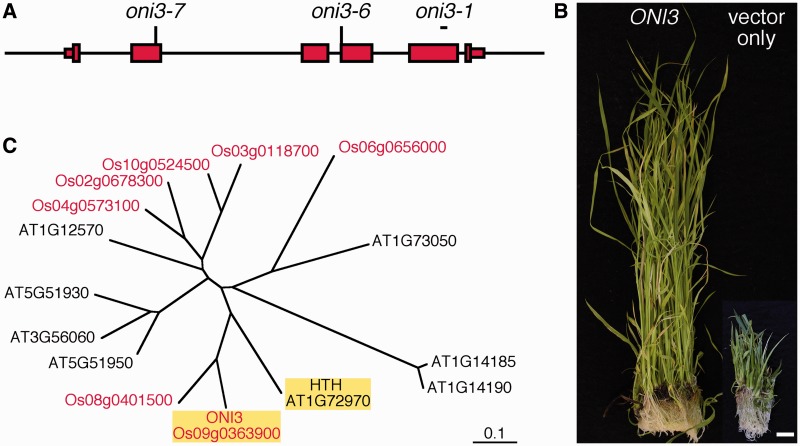
Fig. 5.
Cloning of ONI3. (A) Genome structure of ONI3. Thin and thick red boxes indicate 5′- and 3′-untranslated regions and coding regions, respectively. The line indicates the 5′ and 3′ upstream regions and introns. Positions of the mutations are shown above the genome structure. oni3-6 and oni3-7 had a nucleotide substitution at the indicated position, and oni3-1 had a deletion in the region shown by a short bar. (B) Complementation of oni3. oni3-6 mutant callus transformed with the wild-type ONI3 genome construct showed regeneration of normal shoots, and oni3-6 mutant callus transformed with an empty vector showed regeneration of mutant shoots. Bar = 1 cm. (C) A phylogenetic tree of ω-alcohol dehydrogenase of rice and Arabidopsis drawn on the basis of entire amino acid sequences. Rice proteins are shown in red, and ONI3 and HTH are highlighted by yellow shading.