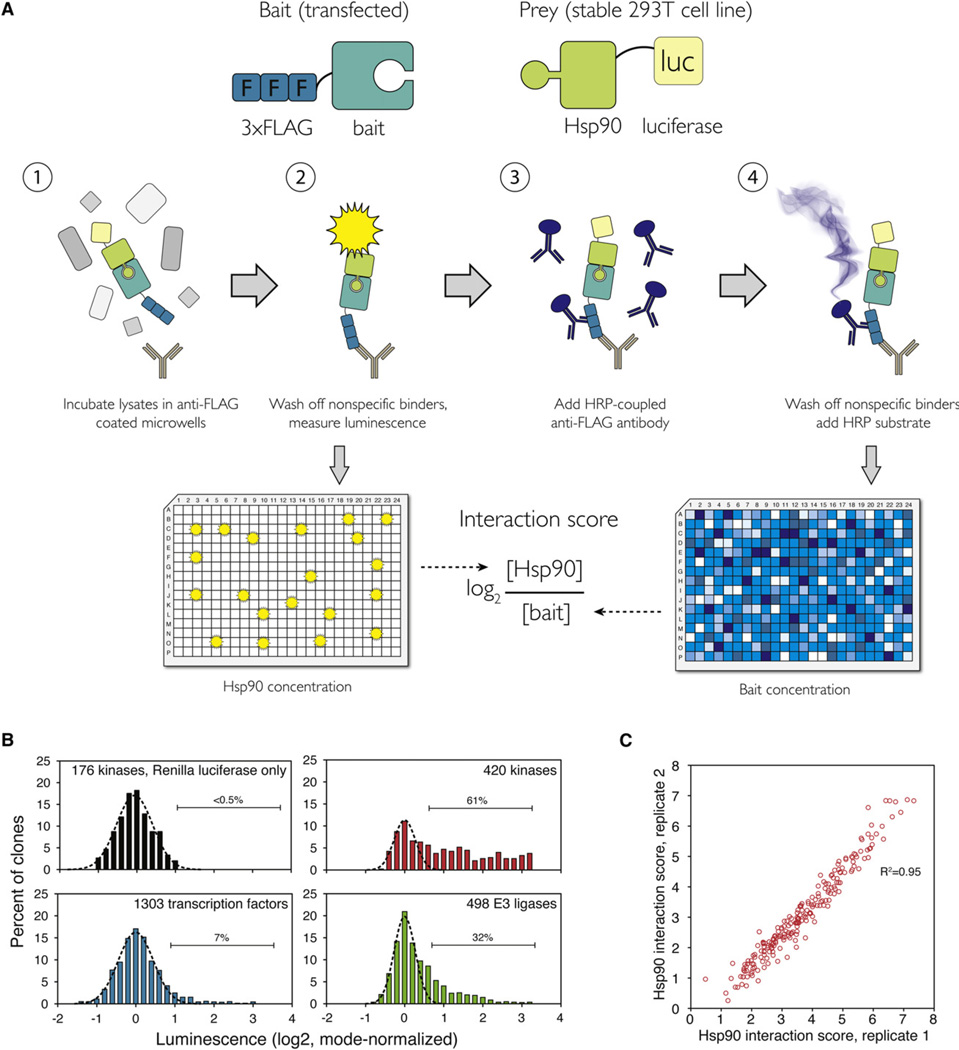
Figure 1. LUMIER with BACON Assay Reveals the Quantitative Nature of HSP90::Client Interactions.
(A) Principle of the assay. 3 × FLAG-tagged bait constructs (putative HSP90 clients) are transfected into a 293T cell line stably expressing Renilla luciferase-tagged HSP90 (prey). Cell lysates are incubated in 384-well plates coated with anti-FLAG antibody. After washing off nonspecific proteins, luminescence is measured. Interaction of HSP90 with the bait can be detected as luminescence. In the second step, the amount of bait is measured with anti-FLAG ELISA. The log2 ratio between bait and prey concentration is the interaction score.
(B) Distribution of luminescence in HSP90 interaction assays with 420 kinase clones (red), 498 E3 ligase clones (green), and 1,093 transcription factor clones (blue). As a control, 176 kinase clones were tested against a cell line expressing Renilla luciferase only (black). Gaussian curve was fitted to each data set to establish a cutoff for true interactions (dashed black line).
(C) Quantitative interaction score was calculated for all 193 kinases that interacted with HSP90. Scatter plot shows the interaction scores from two biological replicates.