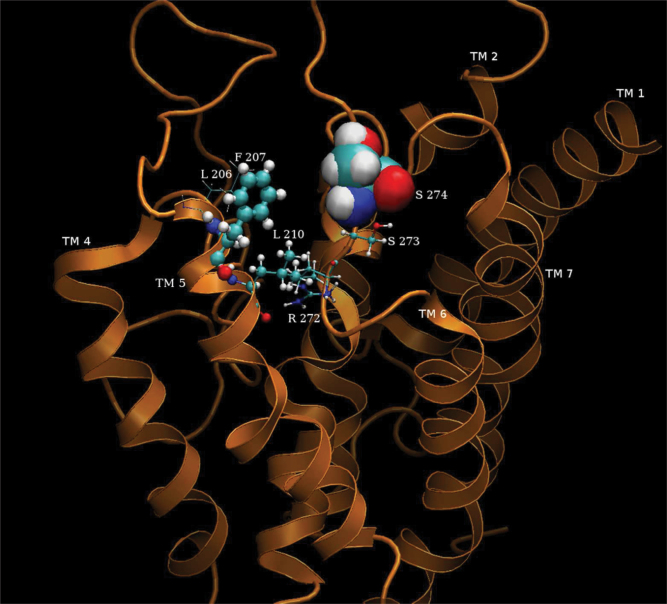
Figure 8.
Results of a 2 ns dynamic simulation of mouse OR S86 and nonanoic acid odor ligand—a strong activator. The close-packed spherical rendering of the side chains of the amino acids are those that are most likely to interact with with 6 Å sphere radius around the odorant ligand over the duration of the simulation. The size of the spheres reflect the aggregated time spent in close contact with the ligand. The top 5 closest contacted amino acid residues in order of time spent in interactions with the odorant ligand are (from highest to lowest): SER274, PHE207, LEU210, SER273, and ARG272.