FIGURE 7.
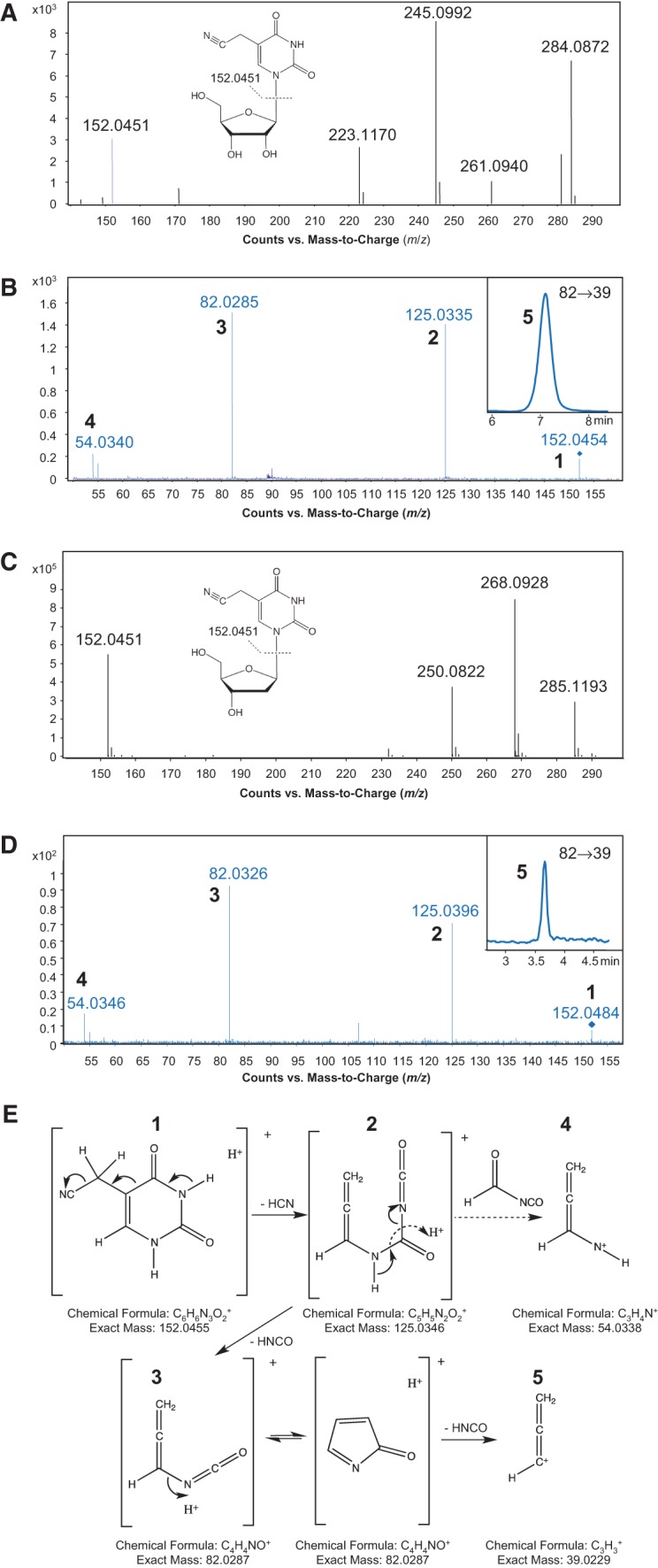
Structural elucidation of 5-cyanomethyl uridine. (A,C) High mass-accuracy mass spectra of the unknown ribonucleoside species (A) found in the purified H. marismortui tRNA2Ile U34 mutant and the 5-cyanomethyl-2′-deoxyuridine synthetic standard (C). The parent ions have m/z values of 284.0872 (A) and 268.0928 (C), and the deglycosylated base ions have identical m/z values of 152.0451 (A,C). (B,D) Pseudo-MS3 mass spectra of the unknown ribonucleoside species (B) and the 5-cyanomethyl-2′-deoxyuridine synthetic standard (D). Following in-source fragmentation of the parent ribonucleoside, the deglycosylated base ion was subjected to CID, with the product ions (numbered 1–5) in close agreement with the theoretical fragmentation pathway shown in panel E. The MRM chromatogram of the m/z 82→39 transition (species 5) is shown in the insets of B and D. (E) Proposed fragmentation pathway for 5-cyanomethyl uracil, with each numbered fragment along with its formula and exact mass.
