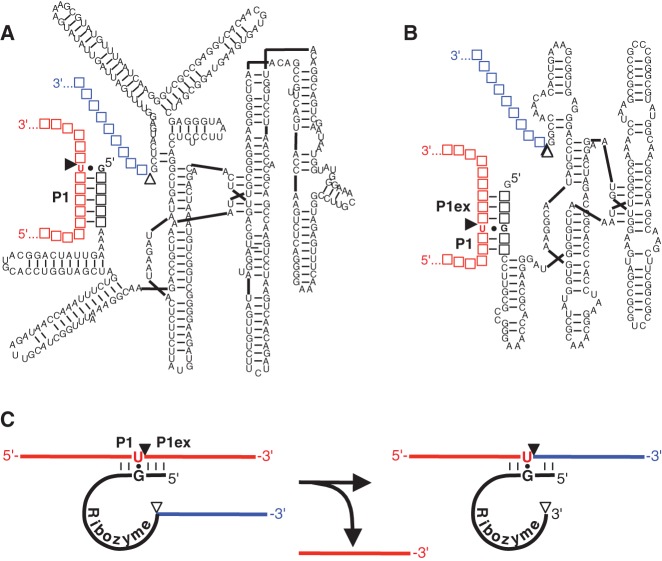
FIGURE 1.
Secondary structure representations of trans-splicing group I intron ribozymes from (A) Tetrahymena and (B) Azoarcus. The 5′ terminus of the trans-splicing ribozymes (black) is truncated at or near the splice site G:U base pair (filled circle) such that the P1 helix (P1) is formed between the ribozyme 5′ terminus and the target site on the substrate (red). This helix is extended on the Azoarcus ribozyme construct by a P1 helix extension (P1ex) to facilitate a similar ribozyme-substrate complex stability as with the Tetrahymena ribozyme. A 5′-terminal guanosine was added for better in vitro transcription yields. Filled triangles indicate the 5′ splice sites, whereas empty triangles denote the 3′ splice sites. Empty squares denote nucleotides that differ between splice sites. (C) Schematic of a trans-splicing reaction. The colors for substrate and ribozyme 3′ exon are as in A and B, whereas sequences are denoted by solid lines (with the exception of the splice site G:U pair), and the ribozyme is denoted by a solid black line. During the trans-splicing reaction, the ribozyme 3′ exon (blue) replaces the 3′ portion of the substrate.