FIGURE 7.
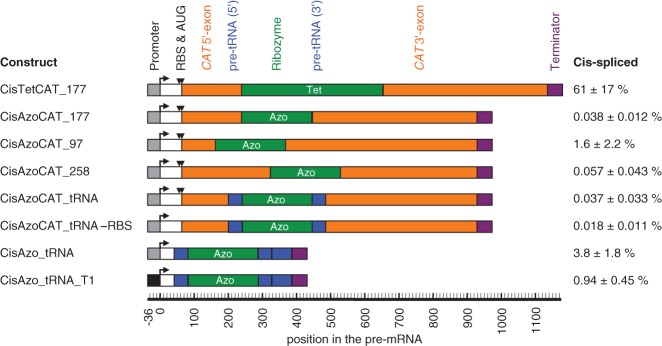
Cis-splicing efficiency of ribozyme constructs in E. coli cells. The name of each construct is given on the left, a schematic representation of the construct is given in the middle, and the observed cis-splicing efficiencies, measured by RT-qPCR, are given on the right. The names of the constructs (“Cis…”) include the ribozyme (“Tet” for Tetrahymena, and “Azo” for Azoarcus), the sequence flanking the ribozyme (CAT, CAT_tRNA, and tRNA), the splice site in the CAT mRNA contexts (177, 97, 258), the mutation inactivation of the ribosomal binding site and the start codon (-RBS) and the use of a different promoter (T1). Note that the three constructs CisAzoCAT_97, CisAzoCAT_177, and CisAzoCAT_258 contained mutations in their 3′ exon immediately downstream from the 3′ splice site to facilitate anticodon stem interactions; the construct CisAzoCAT_97 additionally contained mutations in the P1 loop to generate a P10 helix. For more details on these designs see Supplemental Figure S4. The schematics include the promoter (gray or black boxes with angled arrow), the ribosome-binding site and start codon (RBS and AUG; black triangles), the CAT exons (orange), the ribozymes (green), the flanking tRNAIle sequences (blue), and transcription terminators (purple). A second blue box indicates an extended pre-tRNAIle context. The position of each element in the pre-mRNA is given on a scale below the schematics, relative to the transcription start site. Note that the numbering of splice sites (97, 177, 258) was from the AUG start codon instead. The percentage of cis-spliced mRNA is given on the right, with errors as standard errors from three biological replicates.
