Figure 2.
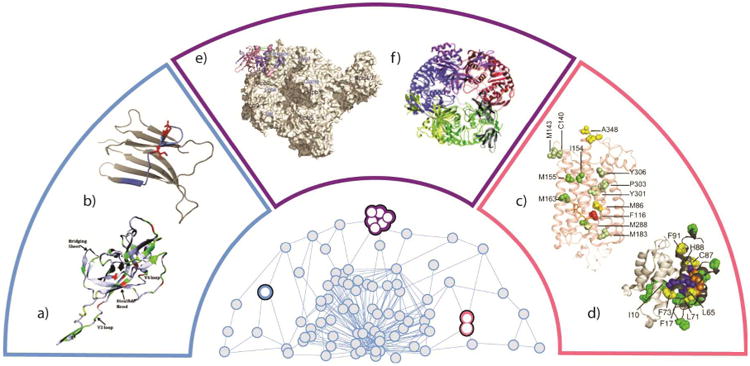
Several MS datasets from the literature are highlighted, and illustrated in context relative to how they might be used in a broader determination of a three-dimensional protein interactome model. These include studies of monomeric protein structure a) gp120-based antigen [70] and b) β-immuoglobulin [51]. Binary protein or protein-ligand systems can also be studied by MS, as in the cases of c) the GPCR-rodopsin complex [71] and d) the S100A11 dimer [69]. Finally, the structure of multiprotein systems can be studied by MS as in the cases of e) the RNA polymerase II [99] and f) the yeast exosome [182]. Each pair of data is contained within color-coded regions that correspond to the same shaded area of the 3D protein interactome cartoon at the center of the figure.
