Abstract
The receptor for advanced glycation end-products (RAGE) is an oncogenic trans-membranous receptor, which is overexpressed in multiple human cancers. However, the role of RAGE in gastric cancer is still elusive. In this study, we investigated the expression and molecular mechanisms of RAGE in gastric cancer cells. Forty cases of gastric cancer and corresponding adjacent non-cancerous tissues (ANCT) were collected, and the expression of RAGE was assessed using immunohistochemistry (IHC) in biopsy samples. Furthermore, RAGE signaling was blocked by constructed recombinant small hairpin RNA lentiviral vector (Lv-shRAGE) used to transfect into human gastric cancer SGC-7901 cells. The expression of AKT, proliferating cell nuclear antigen (PCNA) and matrix metallopeptidase-2 (MMP-2) was detected by Real-time PCR and Western blot assays. Cell proliferative activities and invasive capability were respectively determined by MTT and Transwell assays. Cell apoptosis and cycle distribution were analyzed by flow cytometry. As a consequence, RAGE was found highly expressed in cancer tissues compared with the ANCT (70.0% vs 45.0%, P=0.039), and correlated with lymph node metastases (P=0.026). Knockdown of RAGE reduced cell proliferation and invasion of gastric cancer with decreased expression of AKT, PCNA and MMP-2, and induced cell apoptosis and cycle arrest. Altogether, upregulation of RAGE expression is associated with lymph node metastases of gastric cancer, and blockade of RAGE signaling suppresses growth and invasion of gastric cancer cells through AKT pathway, suggesting that RAGE may represent a potential therapeutic target for this aggressive malignancy.
Key words: RAGE, gastric cancer, growth, invasion
Introduction
Gastric cancer is one of the most common malignancies worldwide, with an estimated 934,000 cases reported globally in 2011, and is the second most common cause of death from cancer.1 Gastric cancer is also a genetic disease developing from a multi-step process. Single or multiple mutations in genes related to growth control, invasion and metastasis form the molecular genetic basis of malignant transformation and tumor progression.2 Thus, identification of predictive biomarkers remains a fundamental challenge in the treatment of patients with gastric cancer.
Receptor for advanced glycation end products (RAGE) is expressed in a range of cell types. The variety of ligands allows RAGE to be implicated in a wide spectrum of pathological conditions such as inflammation and cancer.3 Some studies have shown that, RAGE exhibits high level of expression losing its specific granular pattern characteristic for the primary tumors.4 Co-expression of RAGE and the ligand amphoterin is closely associated with the invasion and metastasis of colorectal cancer.5 The AGE-RAGE interaction plays an important role in the development of prostate cancer, and inhibition of this interaction has potential as a new molecular target for cancer therapy or prevention,6 In addition, RAGE expression is related with the angiogenesis7 and tumor metastasis in oral squamous cell carcinoma (OSCC), and may be an independent prognostic factor for disease recurrence and prognosis in OSCC patients.8 Moreover, the genetic polymorphism of RAGE gene can be used as a genetic marker to screen for patients sensitive to thermotherapy and to predict the prognosis of non-small cell lung cancer (NSCLC).9
However, few studies have indicated that, RAGE expression is strongly reduced at the mRNA and protein levels in NSCLC, and overexpression of RAGE diminishes tumor growth, while downregulation of RAGE supports NSCLC and may be considered as a critical step in tissue reorganization and the formation of NSCLC.10 RAGE expression is negatively correlated with depth of invasion in esophageal squamous cell carcinoma, and tumors with positive RAGE expression exhibit better prognosis than those with negative RAGE expression.11 Therefore, in order to expound the role of RAGE in gastric cancer, we investigated the expression of RAGE in human gastric cancer, and the effects of RAGE knockdown on biological behaviors of gastric cancer cells.
Materials and Methods
Materials
Human SGC-7901 gastric cancer cell line used in the experiments was from Institute of Biochemistry and Cell Biology (Shanghai, China). Lv-shRAGE, negative control vector and virion-packaging elements were from Genechem (Shanghai, China). The primers of RAGE and AKT were synthesized by Applied Biosystems (Carlsbad, CA, USA). All antibodies were from Santa Cruz Biotechnology (Dallas, TX, USA).
Drugs and reagents
Dulbecco’s Modified Eagle medium (DMEM) and fetal bovine serum (FBS) were from Thermo Fisher Scientific Inc. (Waltham, MA, USA); TRIzol Reagent and Lipofectamine 2000 were from Invitrogen (Carlsbad, CA, USA); M-MLV Reverse Transcriptase was from Promega (Madison, WI, USA); SYBR Green Master Mixture was from Takara (Otsu, Japan). ECL-PLUS/Kit was from GE Healthcare (Piscataway, NJ, USA).
Clinical samples and data
Tissue microarray was prepared for immunohistochemical test. Human gastric cancer tissues and the corresponding ANCT were obtained from biopsy prior to chemotherapy in a total of 40 consecutive cases of gastric cancer admitted in our hospital from January 2008 to December 2012. The study was approved by Medical Ethics Committee of Xinjiang Medical University, and written informed consent was obtained from the patients or their parents before sample collection. Two pathologists reviewed all cases.
Tissue microarray
The advanced tissue arrayer (ATA-100, Chemicon International, Tamecula, CA, USA) was used to create holes in a recipient paraffin block and to acquire cylindrical core tissue biopsies with a diameter of 1 mm from the specific areas of the donor block. The tissue core biopsies were transferred to the recipient paraffin block at defined array positions. The tissue microarrays contained tissue samples from 40 formalin-fixed paraffin-embedded cancer specimens with known diagnosis, and corresponding ANCT from these patients. The block was incubated in an oven at 45°C for 20 min to allow complete embedding of the grafted tissue cylinders in the paraffin of the recipient block, and then stored at 4°C until microtome sectioning.
Immunohistochemical staining
Anti-RAGE antibody (Santa Cruz Biotechnology) was used for immunohistochemical (IHC) detection of the expression of RAGE protein in tissue microarrays. Tissue microarray sections were processed for IHC analysis of RAGE protein as follows. IHC examinations were carried out on 3 mm thick sections. For anti-RAGE IHC, unmasking was performed with 10 mM sodium citrate buffer, pH 6.0, at 90°C for 30 min. For anti-RAGE IHC, antigen unmasking was not necessary. Sections were incubated in 0.03% hydrogen peroxide for 10 min at room temperature, to remove endogenous peroxidase activity, and then in blocking serum (0.04% bovine serum albumin, A2153, Sigma-Aldrich, Shanghai, China, and 0.5% normal goat serum X0907, Dako Corporation, Carpinteria, CA, USA, in PBS) for 30 min at room temperature. Anti-RAGE antibody was used at a dilution of 1:200. The antibody was incubated overnight at 4°C. Sections were then washed three times for 5 min in PBS. Non-specific staining was blocked with 0.5% casein and 5% normal serum for 30 min at room temperature. Finally, staining was developed with diaminobenzidine substrate and sections were counterstained with hematoxylin. PBS replaced RAGE antibody in negative controls.
Quantification of protein expression
The expression of RAGE was semiquantitatively estimated as the total immunostaining scores, which were calculated as the product of a proportion score and an intensity score. The proportion and intensity of the staining was evaluated independently by two observers. The proportion score reflected the fraction of positive staining cells (score 0, <5%; score 1, 5%-10%; score 2, 10%-50%; score 3, 50%-75%; score 4, >75%), and the intensity score represented the staining intensity (score 0, no staining; score 1, weak positive; score 2, moderate positive; score 3, strong positive). Finally, a total expression score was given ranging from 0 to 12. Based on the analysis in advance, RAGE was regarded as negative expression in gastric cancer tissues if the score <2, and positive expression if the score ≥2.
Cell culture and infection
SGC-7901 gastric cancer cells were cultured in DMEM medium supplemented with 10% heat-inactivated FBS, 100 U/mL of penicillin and 100 μg/mL of streptomycin. They were all placed in a humidified atmosphere containing 5% CO2 at 37°C. On the day of transduction, SGC-7901 cells were replated at 5×104 cells/well in 24-well plates containing serum-free growth medium with polybrene (5 mg/mL). When reached 50% confluence, cells were transfected with recombinant Lv-shRAGE or control virus at the optimal MOI (multiplicity of infection) of 50, and cultured at 37°C and 5% CO2 for 4 h. Then supernatant was discarded and serum containing growth medium was added. At 4 days of post-transduction, transduction efficiency was measured by the frequency of green fluorescent protein (GFP)-positive cells. Positive stable transfectants were selected and expanded for further study. The clone in which the Lv-shRAGE transfected was named as Lv-shRAGE group, and the negative control vectors transfected was named as NC group.
Quantitative real-time PCR
To quantitatively determine the mRNA expression levels of RAGE and AKT in SGC-7901 cell line, real-time PCR was used. Total RNA of each clone was extracted with TRIzol according to the manufacturer’s protocol. Reverse-transcription was carried out using M-MLV and cDNA amplification was carried out using SYBR Green Master Mix kit according to the manufacturer’s protocol. Target genes were amplified using specific oligonucleotide primer and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as an endogenous control. The PCR primer sequences were as follows: RAGE, 5’-GTCATGGAACTGCCCAAACT-3’ and 5’-TCCTTCTGCGGATCTGTCTT-3’; AKT, 5’-TAGGTGTGGATGGTTGCAGA-3’ and 5’-CTCCCACAGACCCAGAACAT-3’; GAPDH, 5’-CAACGAATTTGGCTACAGCA-3’ and 5’-AGGGGTCTACATGGCAACTG-3’. Data were analyzed using the comparative Ct method (2-ΔCt). Three separate experiments were performed for each clone.
Western blot assay
SGC-7901 cells were harvested and extracted using lysis buffer (Tris-HCl, SDS, Mercaptoethanol, Glycerol). Cell extracts were boiled for 5 min in loading buffer and then equal amount of cell extracts were separated on 15% SDS-PAGE gels. Separated protein bands were transferred into polyvinylidene fluoride (PVDF) membranes and the membranes were blocked in 5% skim milk powder. The primary antibodies against RAGE, AKT, PCNA and MMP-2 were diluted according to the instructions of antibodies and incubated overnight at 4°C. Then, horseradish peroxidase-linked secondary antibodies were added at a dilution ratio of 1:1000, and incubated at room temperature for 2 h. The membranes were washed with PBS for three times and the immunoreactive bands were visualized using ECL-PLUS/Kit according to the kit’s instruction. The relative protein level in different groups was normalized to β-actin concentration. Three separate experiments were performed for each clone.
Cell proliferation assay
Cell proliferation was analyzed with the MTT assay. Briefly, cells infected with Lv-shRAGE were incubated in 96-well-plates at a density of 1×105 cells per well with DEME medium supplemented with 10% FBS. Cells were treated with 20 μL MTT dye at 0, 24 h, 48 h and 72 h, and then incubated with 150 μL of DMSO for 5 min. The color reaction was measured at 570 nm with enzyme immunoassay analyzer (Bio-Rad Laboratories, Hercules, CA, USA). The proliferation activity was calculated for each clone.
Transwell invasion assay
Transwell filters were coated with matrigel (3.9 mg/μL, 60-80 μL) on the upper surface of a polycarbonic membrane (diameter 6.5 mm, pore size 8 mm). After incubating at 37°C for 30 min, the matrigel solidified and served as the extracellular matrix for analysis of tumor cell invasion. Harvested cells (1×105) in 100 μL of serum free DMEM were added into the upper compartment of the chamber. A total of 200 μL conditioned medium derived from NIH3T3 cells was used as a source of chemoattractant, and was placed in the bottom compartment of the chamber. After 24 h incubation at 37°C with 5% CO2, the medium was removed from the upper chamber. The non-invaded cells on the upper side of the chamber were scraped off with a cotton swab. The cells that had migrated from the matrigel into the pores of the inserted filter were fixed with 100% methanol, stained with Hematoxylin, and mounted and dried at 80°C for 30 min. The number of cells invading through the matrigel was counted in three randomly selected visual fields from the central and peripheral portion of the filter using an inverted microscope (200× magnification). Each assay was repeated three times.
Cell apoptosis analysis
To detect cell apoptosis, SGC-7901 cells were trypsinized, washed with cold PBS and resuspended in binding buffer according to the instruction of the apoptosis kit. FITC-AnnexinV and PI were added to the fixed cells for 20 min in darkness at room temperature. Then, Annexin V binding buffer was added to the mixture before the fluorescence was measured on FAC sort flow cytometer. The cell apoptosis was analyzed using the Cell Quest software (Becton Dickinson, Franklin Lakes, NJ, USA). Three separate experiments were performed for each clone.
Cell cycle analysis
To detect cell cycle variation, SGC-7901 cells were trypsinized, washed by PBS and fixed with 80% cold ethanol overnight at -20°C. After PBS washing, the fixed cells were stained with PI in the presence of RNase A for 30 min at room temperature in darkness. Each sample was filtered through a 50 μm nylon filter to obtain single-cell suspension. The samples were then analyzed on FACsort flow cytometer (Becton Dickinson, Mountain View, CA, USA). ModFit3.0 software (Verity Software House, Topsham, ME, USA) was used for cell cycle analysis. Three separate experiments were performed for each clone.
Statistical analysis
SPSS 20.0 was used for the statistical analysis. Kruskal-Wallis H test and Chi-square test were used to analyze the expression rate in all groups. One-way analysis of variance (ANOVA) was used to analyze the differences between groups. The LSD method of multiple comparisons was used when the probability for ANOVA was statistically significant. Statistical significance was set at P<0.05.
Results
Expression of RAGE protein in human gastric cancer
The expression of RAGE protein was evaluated using IHC staining. The positive expression of RAGE protein was detected in the cytoplasm of gastric cancer and ANCT cells (Figure 1). The positive rates of RAGE expression were examined in 70.0% (28/40) of the gastric cancer tissues, and 45.0% (18/40) in a small fraction of ANCT. There was a significant difference between them (P=0.039).
Figure 1.
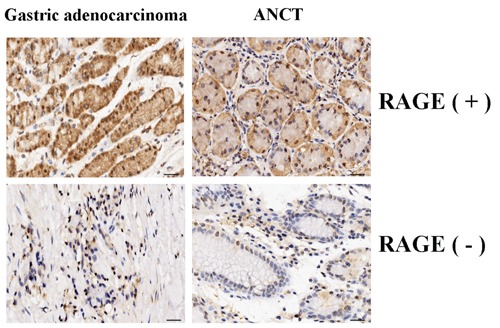
The expression of RAGE protein in human gastric cancer (200×). The expression of RAGE protein was evaluated using immunohistochemical staining. The positive expression of RAGE protein was detected in the cytoplasm of gastric cancer cells and was increased in gastric cancer tissues compared with the adjacent non-cancerous tissues (ANCT). Scale bars: 75 μm.
Correlation of RAGE expression with clinicopathologic characteristics of gastric adenocarcinoma patients
The correlation of RAGE expression with various clinicopathologic characteristics was analyzed. As shown in Table 1, RAGE expression did not associate with the age, gender, tumor size of the gastric adenocarcinoma (GAC) patients (each P>0.05). Up-regulation of RAGE expression did not associate with T stage and N stage (P=0.620; P=0.593). Moreover, positive expression of RAGE was correlated with lymph nodes metastases of the tumors (P=0.026).
Table 1.
Correlation of RAGE expression with the clinicopathologic characteristics of gastric adenocarcinoma patients.
| Variables | Cases | RAGE expression | P | |
|---|---|---|---|---|
| (n) | - | + | ||
| 40 | 12 | 28 | ||
| Age (years) | ||||
| ≥55 | 13 | 3 | 10 | |
| >55 | 27 | 9 | 18 | 0.513 |
| Sex | ||||
| Male | 29 | 8 | 21 | |
| Female | 11 | 4 | 7 | 0.593 |
| Tumor size (cm) | ||||
| ≥3.5 | 8 | 2 | 6 | |
| >3.5 | 32 | 10 | 22 | 0.733 |
| T stage | ||||
| T1 | 7 | 1 | 6 | |
| T2 | 17 | 4 | 13 | |
| T3 | 16 | 7 | 9 | 0.620 |
| N stage | ||||
| N0+N1 | 11 | 4 | ||
| N2+N3 | 29 | 8 | 21 | 0.593 |
| Lymph node metastases | ||||
| Negative | 16 | 8 | 8 | |
| Positive | 24 | 4 | 20 | 0.026 |
Effect of RAGE knockdown on AKT expression
In pilot studies, the infection efficiency of Lv-shRAGE vector (MOI=50) was greater than 70.0% as observed by fluorescent microscopy in SGC-7901 cells (Figure 2). Then, the effect of RAGE knockdown on AKT expression was identified by Real-time PCR and Western blot assays in gastric cancer cells. Real-time PCR showed the lower mRNA expression levels of RAGE and AKT in Lv-shRAGE group than the NC group (each **P<0.01) (Figure 3A). Consistent with this result, the protein expression levels of RAGE and AKT, indicated by Western blot assay were remarkably down-regulated in Lv-shRAGE group in comparison with the NC group (each **P<0.01) (Figure 3B).
Figure 2.
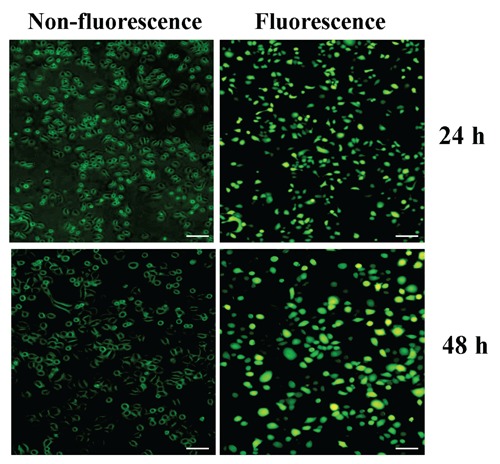
The infection efficiency of Lv-shRAGE vector in gastric cancer cells. In pilot studies, the infection efficiency of Lv-shRAGE vector (multiplicity of infection, MOI=50) was greater than 70.0% as observed by fluorescent microscopy in Lv-shRAGE-transfected SGC-7901 cells. Scale bars: 75 μm.
Figure 3.
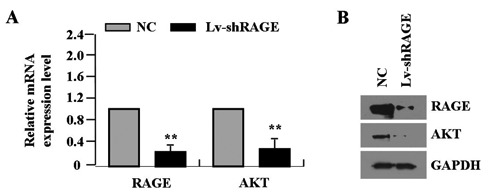
Effect of RAGE knockdown on AKT expression. The effect of RAGE knockdown on AKT expression was identified by real-time PCR (A) and Western blot assays (B) in gastric cancer cells, which indicated the lower expression levels of RAGE and AKT in Lv-shRAGE group than those in NC group in gastric cancer SGC-7901 cells (each **P<0.01).
Effect of RAGE knockdown on cell proliferation
Deregulated cell proliferation is a hallmark of cancer. In order to examine the effect of RAGE knockdown on cell growth, we investigated the proliferative activities of gastric cancer cells by MTT assay. Knockdown of RAGE could significantly diminish the proliferative activities of SGC-7901 cells in a time-dependent manner (**P<0.01) (Figure 4A). In addition, we also detected the expression of PCNA by Real-time PCR and Western blot assays to determine whether knockdown of RAGE suppressed the endogenous PCNA expression. The results showed that, the amount of PCNA was significantly decreased in Lv-shRAGE group compared with the NC group (**P<0.01) (Figure 4 B,C), suggesting that knockdown of RAGE might inhibit gastric cancer cell proliferation through down-regulation of the PCNA expression.
Figure 4.
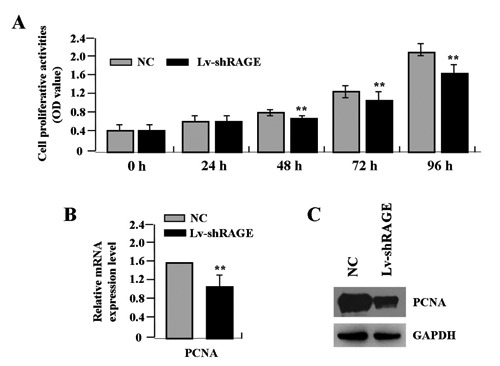
Effect of RAGE knockdown on cell proliferation. A) The proliferative activities of gastric cancer SGC-7901 cells were assessed by MTT assay, indicating that knockdown of RAGE significantly diminished the proliferative activities of SGC-7901 cells in a time-dependent manner (**P<0.01). The amount of PCNA, indicated by real-time (B) and Western blot assays (C), was significantly decreased in Lv-shRAGE group compared with that in NC group (**P<0.01).
Effect of RAGE knockdown on cell invasion
To determine the effect of RAGE knockdown on cell invasion, Transwell assay was carried out. The invasive potential was determined on the basis of the ability of cells to invade a matrix barrier containing laminin and type IV collagen, the major components of the basement membrane. Representative micrographs of Transwell filters can be seen in Figure 5A. The invasive potential of gastric cancer cells was significantly weakened in Lv-shRAGE group than the NC group (**P<0.01) (Figure 5B). In addition, the endogenous expression of MMP-2, indicated by real-time PCR and Western blot assays, was significantly decreased in Lv-shRAGE group compared with the NC group (**P<0.01) (Figure 5 C,D), indicating that knockdown of RAGE might inhibit invasive potential of gastric cancer cells through down-regulation of MMP-2 expression.
Figure 5.
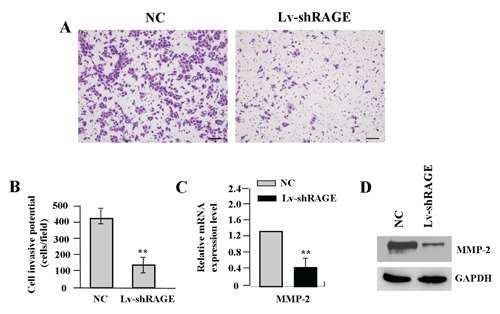
Effect of RAGE knockdown on cell invasion. A) The effect of RAGE knockdown on cell invasion was investigated by Transwell assay. The invasive potential was determined on the basis of the ability of cells to invade a matrix barrier containing laminin and type IV collagen, the major components of the basement membrane. B) The invasive potential of gastric cancer cells was significantly weakened in Lv-shRAGE group compared with that in NC group in SGC-7901 cells (**P<0.01). The amount of MMP-2, indicated by Real-time (C) and Western blot assays (D), was significantly decreased in Lv-shRAGE group compared with that in NC group (**P<0.01). Scale bars: A, 75 μm.
Effect of RAGE knockdown on cell apoptosis and cycle distribution
To determine the effects of RAGE knockdown on apoptosis and cycle distribution in gastric cancer cells (SGC-7901), flow cytometric analysis was performed. The apoptotic indexes of gastric cancer cells in Lv-shRAGE group were markedly higher than the NC group (**P<0.01) (Figure 6A). Cell cycle kinetics showed that, the G0/G1 phase fraction was increased, while S phase fraction was decreased, and cell cycle was arrested in G0/G1 phase in Lv-shRAGE group compared to the NC group (Figure 6B).
Figure 6.
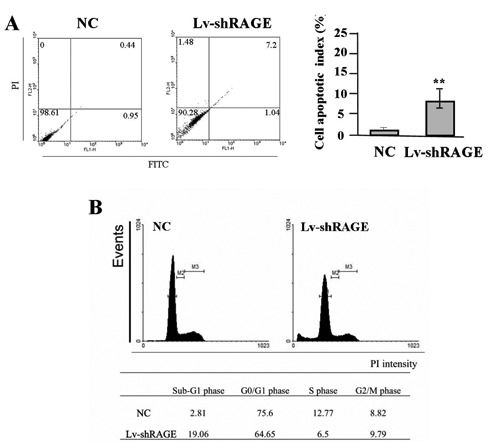
Effect of RAGE knockdown on cell apoptosis and cycle distribution. A) The apoptotic indexes of gastric cancer SGC-7901 cells were markedly higher in Lv-shRAGE group than those in NC group (**P<0.01). B) Cell cycle kinetics showed that, the G0/G1 phase fraction was increased, while S phase fraction was decreased, and cell cycle was arrested in G0/G1 phase in Lv-shRAGE group compared to the NC group in SGC-7901 cells.
Discussion
Gastric cancer is a multistep process that is regulated by intrinsic and extrinsic cellular signals. Extrinsic factors include molecular patterns that are derived from either pathogens or cellular damage, which can promote tumourigenesis. RAGE is a pattern recognition receptor that binds multiple ligands derived from a damaged cell environment, and plays a critical role in promoting the intestinal tumourigenesis.12 RAGE is also an important inflammatory mediator that modulates crosstalk between survival pathways and autophagy in tumor cells. It sustains autophagy and limits apoptosis promoting tumor survival.13 RAGE contributes to formation of pancreatic ductal adenocarcinoma,14 and is permissive for early pancreatic carcinogenesis.15 Therefore, targeted inhibition of RAGE or its ligands may serve as novel targets to enhance current cancer therapies.13
The present study indicated that, RAGE was highly expressed in gastric cancer, but the correlation of RAGE expression with the clinical characteristics of patients with gastric cancer was not further analyzed due to the insufficient clinical data. However, Kuniyasu et al. have reported that, RAGE expression is closely associated with the invasion and metastasis with patients with gastric cancer,16 which provides us an experimental basis for the functional study of RAGE in gastric cancer. Interestingly, Pusterla et al. have identified the novel function of RAGE in regulating oval cell activation and tumor development in inflammation-associated liver carcinogenesis.17 The HMGB1/RAGE inflammatory pathway promotes tumor growth by regulating mitochondrial bioenergetics,18 suggesting that RAGE may represent an important target for the treatment of cancer.
Up to now, the function of RAGE in gastric cancer is unclear, but some studies revealed the role of RAGE in other cancers, in which blockade of RAGE decreased growth and metastases of both implanted tumors and tumors developing spontaneously in susceptible mice.19 Targeting RAGE decreases proliferation in breast cancer cells,20 induces cell apoptosis and inhibits prostate cancer growth.21 Loss of RAGE function also inhibits the angiogenesis and progression of colorectal cancer,22 but prolongs the survival in pancreatic cancer.23 In order to confirm the function of RAGE in gastric cancer, the present study indicated that knockdown of RAGE expression suppressed the growth and invasion, and induced cell apoptosis and cycle arrest in gastric cancer cells. Further work is needed to target RAGE for possible early intervention and prophylaxis in patients at risk for developing cancer. Our previous studies have found that, ethyl pyruvate (EP) can inhibit growth and metastasis of gastric cancer cells via regulation of the HMGB1-RAGE pathway, suggesting that inhibition of RAGE by EP may play a critical role in the treatment of gastric cancer in conjunction with other therapeutic agents.24,25
AKT is overexpressed in many tumor cell lines and in some human tumors including gastric cancer and plays a critical role in tumor growth and metastasis.26,27 PCNA is a nuclear protein that is expressed in proliferating cells, may be required for maintaining cell proliferation, and used as a marker for cancer cell proliferation.28 MMP-2, expressed on the tumor cell surface, is an important molecule involved in tumor metastasis, activates pro-MMP to exacerbate the malignancy, and is considered a powerful indicator of distant metastasis of cancer.29 AKT signaling can promote tumor growth and invasion through regulation of the expression of PCNA and MMP-2.30 Moreover, RAGE leads to the stimulation of the AKT pathway in carcinoma cells, thus enhancing their metastatic capacity.31 In the present study, our finding showed that knockdown of RAGE downregulated the expression of AKT, PCNA and MMP-2, and suppressed the bioactivities of gastric cancer cells, suggesting that RAGE may be involved in the development and progression of gastric cancer via AKT-mediated regulation of the PCNA and MMP-2 expression.
In conclusion, our findings indicate that upregulation of RAGE expression is associated with lymph node metastases of gastric cancer, and blockade of RAGE signaling suppresses growth and invasion of gastric cancer cells through AKT pathway, suggesting that RAGE may represent a potential therapeutic target for this aggressive malignancy.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011;61:69-90 [DOI] [PubMed] [Google Scholar]
- 2.Tajima Y, Yamazaki K, Makino R, Nishino N, Aoki S, Kato M, et al. Gastric and intestinal phenotypic marker expression in early differentiated-type tumors of the stomach: clinicopathologic significance and genetic background. Clin Cancer Res 2006;12:6469-79 [DOI] [PubMed] [Google Scholar]
- 3.Alexiou P, Chatzopoulou M, Pegklidou K, Demopoulos VJ. RAGE: a multi-ligand receptor unveiling novel insights in health and disease. Curr Med Chem 2010;17:2232-52 [DOI] [PubMed] [Google Scholar]
- 4.Kostova N, Zlateva S, Ugrinova I, Pasheva E. The expression of HMGB1 protein and its receptor RAGE in human malignant tumors. Mol Cell Biochem 2010;337:251-8 [DOI] [PubMed] [Google Scholar]
- 5.Kuniyasu H, Chihara Y, Takahashi T. Co-expression of receptor for advanced glycation end products and the ligand amphoterin associates closely with metastasis of colorectal cancer. Oncol Rep 2003;10:445-8 [PubMed] [Google Scholar]
- 6.Ishiguro H, Nakaigawa N, Miyoshi Y, Fujinami K, Kubota Y, Uemura H. Receptor for advanced glycation end products (RAGE) and its ligand, amphoterin are overexpressed and associated with prostate cancer development. Prostate 2005;64:92-100 [DOI] [PubMed] [Google Scholar]
- 7.Sasahira T, Kirita T, Bhawal UK, Ikeda M, Nagasawa A, Yamamoto K, et al. The expression of receptor for advanced glycation end products is associated with angiogenesis in human oral squamous cell carcinoma. Virchows Arch 2007;450:287-95 [DOI] [PubMed] [Google Scholar]
- 8.Sasahira T, Kirita T, Bhawal UK, Yamamoto K, Ohmori H, Fujii K, et al. Receptor for advanced glycation end products (RAGE) is important in the prediction of recurrence in human oral squamous cell carcinoma. Histopathology 2007;51:166-72 [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Cui E, Zeng H, Hua F, Wang B, Mao W, et al. RAGE genetic polymorphisms are associated with risk, chemotherapy response and prognosis in patients with advanced NSCLC. PLoS One 2012;7:e43734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartling B, Hofmann HS, Weigle B, Silber RE, Simm A. Down-regulation of the receptor for advanced glycation end-products (RAGE) supports non-small cell lung carcinoma. Carcinogenesis 2005;26:293-301 [DOI] [PubMed] [Google Scholar]
- 11.Tateno T, Ueno S, Hiwatashi K, Matsumoto M, Okumura H, Setoyama T, et al. Expression of receptor for advanced glycation end products (RAGE) is related to prognosis in patients with esophageal squamous cell carcinoma. Ann Surg Oncol 2009;16:440-6 [DOI] [PubMed] [Google Scholar]
- 12.Heijmans J, Büller NV, Hoff E, Dihal AA, van der Poll T, van Zoelen MA, et al. Rage signalling promotes intestinal tumourigenesis. Oncogene 2013;32:1202-6 [DOI] [PubMed] [Google Scholar]
- 13.Kang R, Tang D, Schapiro NE, Livesey KM, Farkas A, Loughran P, et al. The receptor for advanced glycation end products (RAGE) sustains autophagy and limits apoptosis, promoting pancreatic tumor cell survival. Cell Death Differ 2010;17:666-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang R, Tang D, Lotze MT, Zeh HJ., 3rd AGER/RAGE-mediated autophagy promotes pancreatic tumorigenesis and bioenergetics through the IL6-pSTAT3 pathway. Autophagy 2012;8:989-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang R, Loux T, Tang D, Schapiro NE, Vernon P, Livesey KM, et al. The expression of the receptor for advanced glycation endproducts (RAGE) is permissive for early pancreatic neoplasia. Proc Natl Acad Sci USA 2012;109:7031-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuniyasu H, Oue N, Wakikawa A, Shigeishi H, Matsutani N, Kuraoka K, et al. Expression of receptors for advanced glycation end-products (RAGE) is closely associated with the invasive and metastatic activity of gastric cancer. J Pathol 2002;196:163-70 [DOI] [PubMed] [Google Scholar]
- 17.Pusterla T, Nèmeth J, Stein I, Wiechert L, Knigin D, Marhenke S, et al. Rage is a key regulator of oval cell activation and inflammation-associated liver carcinogenesis in mice. Hepatology 2013;58:363-73 [DOI] [PubMed] [Google Scholar]
- 18.Kang R, Tang D, Schapiro NE, Loux T, Livesey KM, Billiar TR, et al. The HMGB1/RAGE inflammatory pathway promotes pancreatic tumor growth by regulating mitochondrial bioenergetics. Oncogene 2013. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, Qu W, et al. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature 2000;405:354-60 [DOI] [PubMed] [Google Scholar]
- 20.Radia AM, Yaser AM, Ma X, Zhang J, Yang C, Dong Q, et al. Specific siRNA Targeting Receptor for Advanced Glycation End Products (RAGE) Decreases Proliferation in Human Breast Cancer Cell Lines. Int J Mol Sci 2013;14:7959-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elangovan I, Thirugnanam S, Chen A, Zheng G, Bosland MC, Kajdacsy-Balla A, et al. Targeting receptor for advanced glycation end products (RAGE) expression induces apoptosis and inhibits prostate tumor growth. Biochem Biophys Res Commun 2012;417:1133-8 [DOI] [PubMed] [Google Scholar]
- 22.Liang H, Zhong Y, Zhou S, Peng L. Knockdown of RAGE expression inhibits colorectal cancer cell invasion and suppresses angiogenesis in vitro and in vivo. Cancer Lett 2011;313:91-8 [DOI] [PubMed] [Google Scholar]
- 23.DiNorcia J, Lee MK, Moroziewicz DN, Winner M, Suman P, Bao F, et al. RAGE gene deletion inhibits the development and progression of ductal neoplasia and prolongs survival in a murine model of pancreatic cancer. J Gastrointest Surg 2012;16:104-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.J Zhang, J-S Zhu, Z Zhou, W-X Chen, N-W Chen. Inhibitory effects of ethyl pyruvate administration on human gastric cancer growth via regulation of the HMGB1-RAGE and Akt pathways in vitro and in vivo. Oncol Rep 2012;27:1511-19 [DOI] [PubMed] [Google Scholar]
- 25.J Zhang, J-S Zhu, Z Zhou, W-X Chen, N-W Chen. Therapeutic effects of ethyl pyruvate on growth and metastasis in a severe combined immunodeficiency mice orthotopic implantation model. Eur J Inflamm 2012;10:25-32 [Google Scholar]
- 26.Ye B, Jiang LL, Xu HT, Zhou DW, Li ZS. Expression of PI3K/AKT pathway in gastric cancer and its blockade suppresses tumor growth and metastasis. Int J Immunopathol Pharmacol 2012;25:627-36 [DOI] [PubMed] [Google Scholar]
- 27.Matsuoka T, Yashiro M, Nishioka N, Hirakawa K, Olden K, Roberts JD. PI3K/Akt signalling is required for the attachment and spreading and growth in vivo of metastatic scirrhous gastric carcinoma. Br J Cancer 2012;106:1535-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall PA, Levison DA, Woods AL, Yu CC, Kellock DB, Watkins JA, et al. Proliferating cell nuclear antigen (PCNA) immunolocalization in paraffin sections: an index of cell proliferation with evidence of deregulated expression in some neoplasms. J Pathol 1990;162:285-94 [DOI] [PubMed] [Google Scholar]
- 29.Ueda J, Kajita M, Suenaga N, Fujii K, Seiki M. Sequence-specific silencing of MT1-MMP expression suppresses tumor cell migration and invasion: importance of MT1-MMP as a therapeutic target for invasive tumors. Oncogene 2003;22:8716-22 [DOI] [PubMed] [Google Scholar]
- 30.Pu P, Kang C, Li J, Jiang H, Cheng J. The effects of antisense AKT2 RNA on the inhibition of malignant glioma cell growth in vitro and in vivo. J Neurooncol 2006;76:1-11 [DOI] [PubMed] [Google Scholar]
- 31.Tafani M, Schito L, Pellegrini L, Villanova L, Marfe G, Anwar T, et al. Hypoxia-increased RAGE and P2X7R expression regulates tumor cell invasion through phosphorylation of Erk1/2 and Akt and nuclear translocation of NF-{kappa}B. Carcinogenesis 2011;32:1167-75 [DOI] [PubMed] [Google Scholar]


