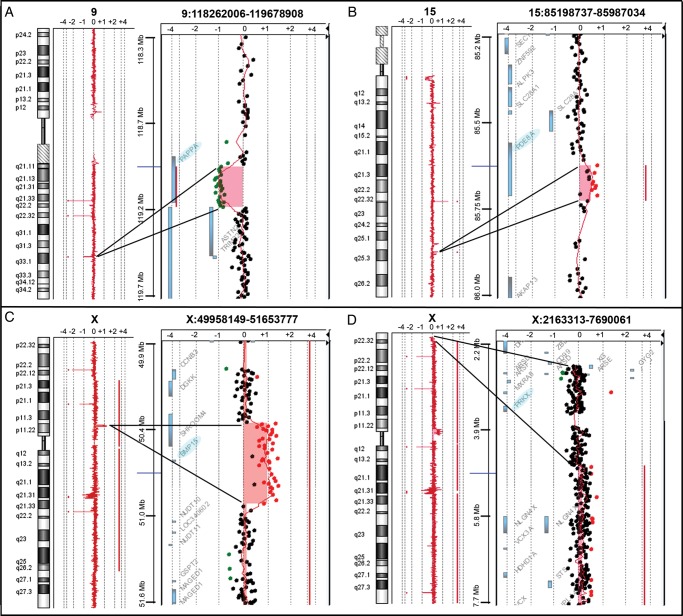
Figure 1.
Detection of copy number variations. High-resolution aCGH analysis 244 K Agilent identified in patient PA5 a deletion of at least 216 kb at 9q33.1 (chr9:118971743-119187538, hg19), which interrupts the PAPPA gene coding region (A), in patient PA4 a duplication of at least 75 kb at 15q25.3 (chr15:85591615-85666309, hg19), which includes part of the PDE8A gene coding region (B), and in patient SM1 a duplication of at least 554 kb at Xp11.22 (chrX:50400649-50955142, hg19,), which contains the entire BMP15 gene (C). In addition, a mosaic deletion of at least 2 Mb at Xp22.33-p22.32 (chrX:2700316-4739185, hg19), which includes the entire PRKX gene, was detected in patient PA4 (D). The most important genes involved in the identified microrearrangements are highlighted in light blue.