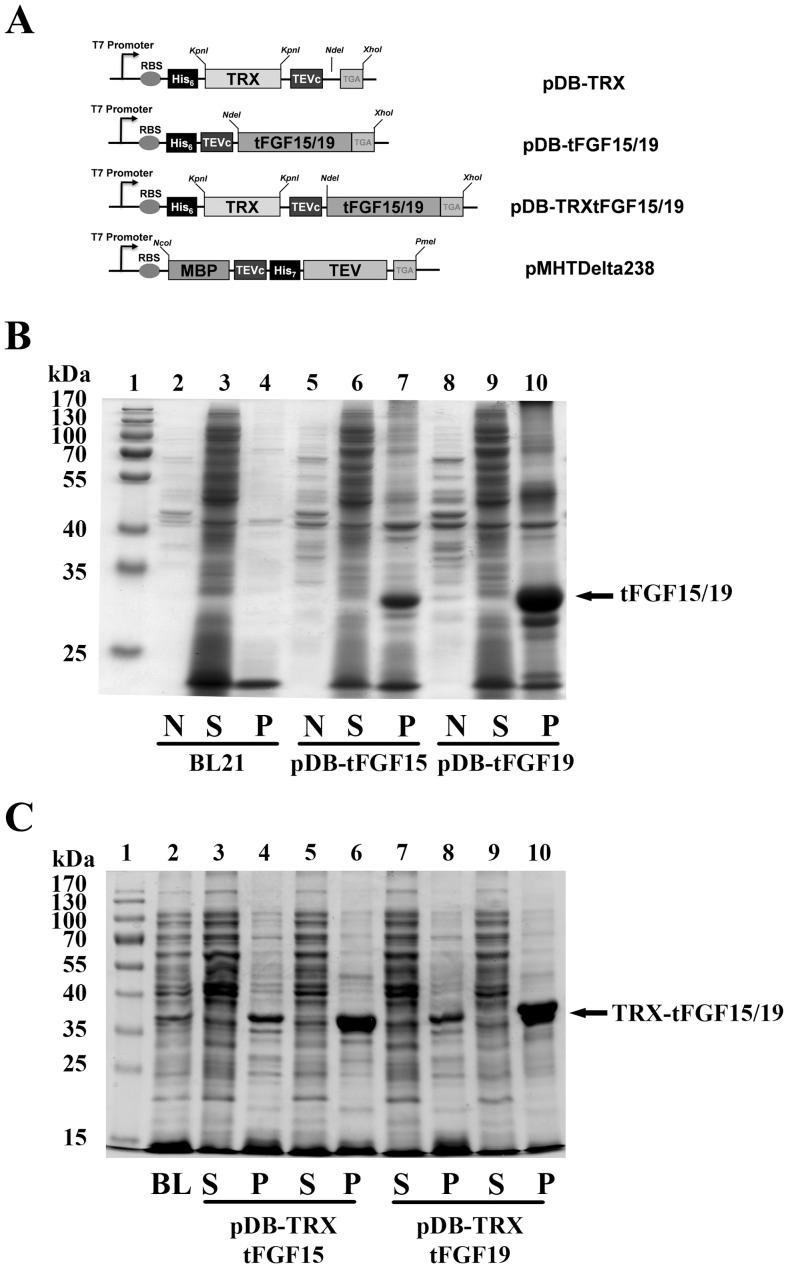
Figure 1. Construction and expression analyses of FGF15 and FGF19 Plasmids.
A): Schematic maps of plasmids. Truncated FGF15 or FGF19 gene (without signal peptide) was located downstream from the T7 promoter, coding in frame with the TRX tag. NdeI and XhoI restriction enzyme sites are used to subclone the PCR fragments. His6 tag for purification and TEV protease site for fusion tag cleavage are indicated in the figure. B): SDS-PAGE analysis of FGF15 and FGF19 in E. coli strain BL21(DE3); non-induced total cell lysates (N), and the soluble (S) and insoluble fractions (P) were prepared as described in Methods. The molecular weight markers from Pierce (Rockford, IL) are also shown in gels. C): SDS-PAGE analysis of TRX-FGF15/19 in BL21(DE3) strain; the soluble (S) and insoluble fractions (P) before (Lane 3, 4, 7, and 8) and after IPTG induction (lane 5, 6, 9 and 10) were determined. Total cell lysates of BL21(DE3) strain (BL) were loaded as the control.