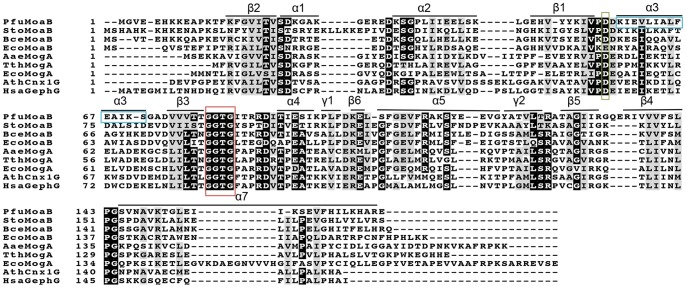
Figure 4. Multiple sequence alignment of MPT-adenylyl-transferases from different organisms.
Corresponding MPT-adenylyl-transferases are abbreviated as follows: PfuMoaB, Pyrococcus furious; StoMoaB, Sulfolobus tokodaii; BceMoaB, Bacillus cereus; EcoMogA and EcoMoaB, Escherichia coli; TthMogA, Thermus thermophilus; AaeMogA, Aquifex aeolicus; Arabidopsis thaliana; HsaGephG, Homo sapiens. Secondary structure elements of PfuMoaB are shown. The conserved MPT-binding motif GGTG is highlighted with a red box, the conserved aspartate residue coordinating Mg2+- ion with a green box [6], residues of PfuMoaB α3-helix with a blue box. Highly conserved residues are depicted in white letters and black background; semi-conserved residues are shadowed in grey. Consensus threshold was set to 0.8. Sequences were aligned with Clustal Omega [62],and modified with BoxShade server (Swiss Institute of Bioinformatics).