Abstract
Background
Ageing affects many components of the immune system, including innate immune cells like monocytes. They are important in the early response to pathogens and for their role to differentiate into macrophages and dendritic cells. Recent studies have revealed significant age-related changes in genomic DNA methylation in peripheral blood mononuclear cells, however information on epigenetic changes in specific leukocyte subsets is still lacking. Here, we aimed to analyse DNA methylation in purified monocyte populations from young and elderly individuals.
Findings
We analysed the methylation changes in monocytes purified from young and elderly individuals using the HumanMethylation450 BeadChip array. Interestingly, we found that among 26 differentially methylated CpG sites, the majority of sites were hypomethylated in elderly individuals. The most hypomethylated CpG sites were located in neuropilin 1 (NRP1; cg24892069) and neurexin 2 (NRXN2; cg27209729) genes, and upstream of miR-29b-2 gene (cg10501210). The age-related hypomethylation of these three sites was confirmed in a separate group of young and elderly individuals.
Conclusions
We identified significant age-related hypomethylation in human purified monocytes at CpG sites within the regions of NRP1, NRXN2 and miR-29b-2 genes.
Keywords: Monocytes, DNA methylation, Ageing
Main text
Innate and adaptive immune responses are affected by ageing. Elderly people have a decreased ability to maintain basic tissue homeostasis, impaired vaccination responses and an increased risk for infectious diseases, particularly influenza virus [1-4]. A diverse range of age-associated changes has been reported in human innate immune cells [3,5], which are important during the early response to pathogens. Monocytes, which are circulating cells that originate from myeloid precursors, are the precursors of tissue macrophages and dendritic cells and constitute an essential part of the innate immune system. Although the number of monocytes does not change significantly during ageing, several functional age-related changes in monocytes, such as altered expression of cytokines, defective Toll-like receptor signalling and a decreased capacity for phagocytosis, have been reported [6]. Monocytes are also involved in the initiation of atherosclerosis on arterial walls and have been linked to a chronic inflamed state (referred to as inflamm-ageing), which is associated with increased cardiovascular and metabolic diseases in elderly individuals [7]. Recent studies have revealed the important role of epigenetic regulation in the development and cell-specific functions of blood cells. Changes in DNA methylation patterns occur gradually throughout an individual’s lifespan [8,9] and may result in the age-related phenotypes of a specific set of genes [8]. The majority of these studies have examined DNA methylation changes in a mixed population of peripheral blood mononuclear cells (PBMCs) without purifying specific subsets of cells. In this study, we aimed to analyse the epigenomic changes in DNA methylation in purified monocyte cell populations from young and elderly individuals.
To study age-related DNA methylation profiles, we isolated monocytes from the peripheral blood of eight young (age range 22–25 years, mean 23.75 years; 4 females and 4 males) and eight elderly healthy volunteers (age range 77–78 years, mean 77.13 years; 4 females and 4 males). A whole genome methylation analysis was performed using the Infinium HumanMethylation450 BeadChip (Illumina Inc.). Altogether, we found 368 CpG sites that were significantly differentially methylated (p < 0.05), of which 26 CpG sites had an absolute β value differences greater than or equal to 0.2 between the young and old individuals (Table 1). Most of the CpG sites, a total of 21 positions, were hypomethylated in the elderly individuals; only five positions were hypermethylated in these individuals. Decreased methylation during the ageing process has been previously described in a study of PBMCs [10]. The most significantly altered sites mapped within the NRP1, NRXN1, RASSF5, OTUD7A and PRM1 genes. The loci that did not reach the 0.2 β-difference threshold but were significantly different (p < 0.05) included two ELOVL2 sites, cg16867657 and cg24724428 (both with β-diff. of 0.17); two FHL2 sites, cg22454769 and cg24079702 (β-diff. of 0.15 and 0.14, respectively); and a PENK site, cg16419235 (β-diff. of 0.08); all these sites are associated with increased methylation in the peripheral blood mononuclear cells of older individuals [9,10].
Table 1.
Differentially methylated sites in young versus old monocyte cell populations
| Target ID | β-difference* | Adjusted p-value | Gene |
|---|---|---|---|
|
cg10501210 |
−0.38 |
0.002884 |
miR-29b-2
*
*
|
|
cg24892069 |
−0.30 |
0.00312 |
NRP1 |
|
cg27209729 |
−0.30 |
0.020571 |
NRXN2 |
| cg11807280 |
−0.27 |
0.013523 |
|
| cg08128734 |
−0.27 |
0.015965 |
RASSF5 |
| cg11693709 |
−0.26 |
0.024853 |
PAK6 |
| cg18826637 |
−0.25 |
0.022927 |
|
| cg00329615 |
−0.25 |
0.012874 |
IGSF11 |
| cg00740914 |
−0.25 |
0.008061 |
|
| cg03873281 |
−0.25 |
0.005647 |
PDLIM4 |
| cg13039251 |
−0.23 |
0.008793 |
PDZD2 |
| cg07583137 |
−0.23 |
0.007904 |
CHMP4C |
| cg12317815 |
−0.22 |
0.014773 |
ASPA |
| cg06781608 |
−0.22 |
0.030063 |
PTPRN2 |
| cg13001142 |
−0.22 |
0.011913 |
STXBP5 |
| cg16932827 |
−0.21 |
0.015062 |
|
| cg19344626 |
−0.21 |
0.010547 |
NWD1 |
| cg14295611 |
−0.21 |
0.04857 |
|
| cg14614643 |
−0.21 |
0.008462 |
|
| cg03915012 |
−0.21 |
0.026415 |
GAK |
| cg03473532 |
−0.20 |
0.035003 |
MKLN1 |
| cg02978201 |
0.47 |
0.015062 |
PRM1 |
| cg04875128 |
0.30 |
0.017558 |
OTUD7A |
| cg21184711 |
0.23 |
0.024671 |
CADPS2 |
| cg20665157 |
0.22 |
0.008629 |
CADPS2 |
| cg19907915 | 0.21 | 0.014934 | IGSF9B |
The three replicated CpG loci are shown in bold.
*A negative β-difference indicates to hypomethylation in the elderly and a positive β-difference indicates to hypermethylation in the elderly population. Only CpG sites with FDR-adjusted p-values less than 0.05 were considered differentially methylated.
**Approximately 1 kb upstream.
To validate our results, we focused our investigation on the three differentially methylated CpG sites with the highest hypomethylation values, cg24892069, cg27209729 and cg10501210. The CpG site cg24892069, which had a very low standard deviation in both age groups (young STDEV: 0.05; old STDEV: 0.06), is located in intron 2 of the neuropilin 1 (NRP1) gene. NRP1 is a cell surface receptor with functional roles in several biological processes, including angiogenesis, immune response and regulation of vascular permeability [11,12], and has also been associated with increased cancer progression [13,14]. NRP1 is expressed in regulatory T cells [15] and is needed for prolonged cellular contact between regulatory T-cells and dendritic cells [16]. Another CpG site, cg27209729, is located in intron 9 of the neurexin 2 (NRXN2) gene. NRXN2 is a member of the neurexin family, which affects synaptic plasticity and cognitive functioning [17], and has been linked to autism spectrum disorders and schizophrenia [18]. The third CpG site, cg10501210, is located in putative regulatory region, approximately 1 kb upstream of the miR-29b-2 gene. miR-29b-2 belongs to the miR-29 family, which is important in thymic involution [19], T cell polarisation [20] and oncogenesis [19,21]. The miR-29b has been shown to target DNA methyltransferases DNMT3A and DNMT3B, and indirectly DNMT1 [22,23], leading to reduction of global methylation and expression of methylation regulated genes.
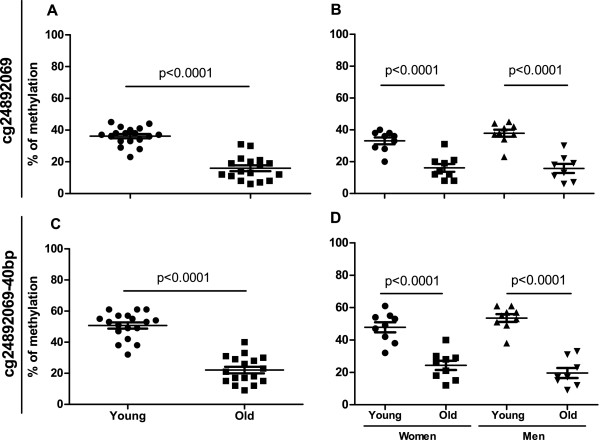
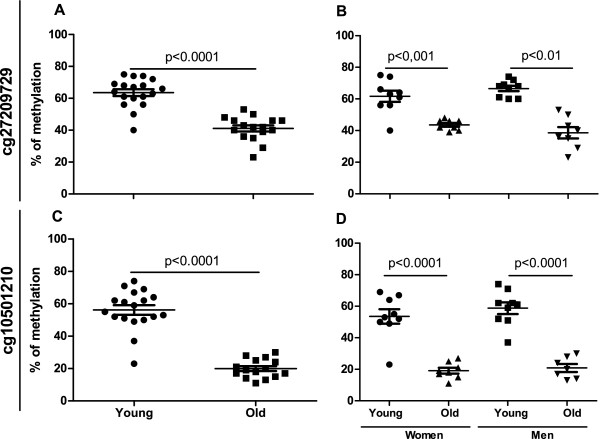
We replicated the array results of the three differentially methylated loci using the EpiTYPER assay (Sequenom Inc.) with a separate set of young and elderly samples. We added to our analyses two sex-matched control age groups, consisting of 10 young (age range 24–28 years, mean 26.4 years; 5 men and 5 females) and 10 elderly (age range 76–84 years, mean 79.4 years; 5 men and 5 females) samples. Using the EpiTYPER assay, we found hypomethylation of the NRP1-associated cg24892069 site in the monocytes of the older individuals, similar to the results from the HumanMethylation450 BeadChip analysis (Figure 1A). We also analysed the methylation differences in men and women separately and observed a significant difference in both gender groups (p < 0.0001) (Figure 1B). To explore this region further, we selected another CpG site, cg24892069-40 bp, which was located 40 bp upstream of the cg24892069 site in the genomic sequence; this site was not included on the methylation BeadChip. We found that the cg24892069-40 bp site had a statistically significant methylation difference between the studied age groups (p < 0.0001) (Figure 1C) and that was observed in both sexes (p < 0.0001) (Figure 1D). The similar DNA methylation pattern of the two CpG sites in close proximity is most likely the result of a shared, differentially methylated, region that is modified from the nearby methyltransferase binding site. We also found significant differences between the age groups at the cg27209729 and cg10501210 sites, located in the NRXN2 gene and upstream of the miR-29b-2 gene, respectively (Figure 2). These CpG sites had statistically significant methylation differences in the combined study group (p < 0.0001) (Figure 2 A&C) as well as in the male and female study groups (p < 0.01 and p < 0.0001, respectively) (Figure 2 B&D).
Figure 1.
Percentage of methylation of CpG sites cg24892069 (NRP1; A & B) and cg24892069-40 bp (NRP1; C & D). The mean (±S.E.M.) methylation difference measured in 18 young and 17 old individuals using the Sequenom’s EPITYPER assay.
Figure 2.
Percentage of methylation of CpG sites cg27209729 (NRXN2; A & B) and cg10501210 (miR-29b-2; C & D). The mean (±S.E.M.) methylation difference measured in 18 young and 15 old individuals using the Sequenom’s EPITYPER assay.
We also evaluated the expression of the three differentially methylated CpG sites in monocytes of young and elderly individuals, but the expression levels NRP1 and NRXN2 genes were under the detection limit of RT-PCR. This is in agreement with our previously published mRNA expression study, where NRP1 was expressed at very low levels in monocytes and demonstrated a significantly increased expression in monocyte-derived dendritic cells and macrophages, whereas NRXN2 expression remained low even after the differentiation to dendritic cells [24]. The mRNA level of miR-29b-2 gene was detectable, however, the expression between young and elderly individuals did not differ significantly (data not shown). As the CpG site cg10501210 is located approximately 1 kb upstream of miR-29b-2 gene, it might not have regulatory effect on miR-29b-2 gene expression.
In conclusion, we were able to identify age-related DNA methylation changes in purified monocytes at immunologically relevant genomic loci. We found that the majority of the altered CpG sites were hypomethylated in the elderly individuals. The top three hypomethylated CpG sites in the elderly were cg24892069, cg27209729 and cg10501210, which are located in or near the NRP1, NRXN2 and miR-29b-2 genes, respectively. Further investigation and a larger sample set are needed to define the functional role and significance of these CpG sites in the ageing process.
Material and methods
Purification of cell populations
The study is approved by Ethics Review Committee on Human Research of the University of Tartu. All of the participants gave written informed consent. Peripheral blood was obtained from healthy donors of Estonian Genome Center of University of Tartu. Peripheral blood mononuclear cells (PBMC) were extracted using a Ficoll-Paque (GE Healthcare) gradient centrifugation. CD14+ monocytes were extracted from PBMCs using microbeads (CD14+ #130-050-201) and AutoMACS technology (Miltenyi Biotec). The purity of monocyte cell population was analysed with FACSCalibur (BD Biosciences) using fluorescence conjugated antibodies against CD14 and CD3 (Miltenyi) to confirm the characteristic phenotype (Additional file 1: Figure S1).
DNA extraction, bisulfite treatment and DNA methylation measurement
Genomic DNA was isolated from cell pellets using QIAmp DNA Micro Kit (Qiagen). DNA concentration was measured with NanoDrop ND-1000 spectrophotometry. Extracted genomic DNA was bisulfite converted using EZ-96 DNA Methylation Kit (Zymo Research Corporation). DNA methylation analysis was performed using Infinium Human Methylation 450 K bead chip technology (Illumina).
Sequenom EpiTYPER assay
The Sequenom EpiTYPER technology was used to validate HumanMethylation450 array data. Samples were prepared using EpiTYPER T Complete Reagent Set (Sequenom) according to manufacturer’s instructions. 25 ng of bisulfite-treated DNA was used as PCR input and CpG methylation was determined by the MassARRAY Analyzer 4 system (Sequenom).
Data analyses
The methylation signals were extracted with the methylation module v1.8.5 of the GenomeStudio v2010.3 software (Illumina Inc.) without background correction and normalisation. Probes with a detection p-value greater than 0.01, located on sex chromosomes or containing SNPs with a minor allele frequency of at least 5% in the Caucasian population according to the Hapmap project (http://hapmap.ncbi.nlm.nih.gov) were filtered out prior further analysis. The signals were corrected and normalised using subset quantile normalisation as described in [25]. For differential methylation analysis, 80% of the least varying probes according to interquartile range across all samples were removed and a linear model was used to assess the differences between two age groups considering arrays on different BeadChips as batches. Methylation sites with a FDR adjusted p-value less than 0.05 were considered differentially methylated. Median difference of beta values greater than 0.2 between groups was considered for selecting methylation sites for further analyses.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TL and PP designed the study and wrote the manuscript; TL purified the cells and carried out the methylation array; LM carried out the validation experiments; SM analysed the data and helped to interpret the data; KK, MA and ML contributed to the design of the study and coordinated the recruitment of the study participants. All authors read and approved the final manuscript.
Supplementary Material
The purification of monocyte cell population. The monocytes were analysed with FACSCalibur (BD Biosciences) using fluorescence conjugated antibodies against CD14 and CD3 (Miltenyi).
Contributor Information
Liina Tserel, Email: liina.tserel@ut.ee.
Maia Limbach, Email: maia.limbach@ut.ee.
Mario Saare, Email: mario.saare@ut.ee.
Kai Kisand, Email: kai.kisand@ut.ee.
Andres Metspalu, Email: andres.metspalu@ut.ee.
Lili Milani, Email: lili.milani@ut.ee.
Pärt Peterson, Email: part.peterson@ut.ee.
Acknowledgements
The study was supported by the Estonian Research Agency grant IUT2-2, the Center of Excellence of Translational Medicine, the Tartu University Development Fund and the Center of Translational Genomics. We would like to thank all of the participants of this study and all of the people who helped to collect and process the material.
References
- Haynes L, Swain SL. Why aging T cells fail: implications for vaccination. Immunity. 2006;24:663–666. doi: 10.1016/j.immuni.2006.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaiou RD, Herndler-Brandstetter D, Grubeck-Loebenstein B. Age-related changes in immunity: implications for vaccination in the elderly. Expert Rev Mol Med. 2007;9:1–17. doi: 10.1017/S1462399407000221. [DOI] [PubMed] [Google Scholar]
- Panda A, Arjona A, Sapey E, Bai F, Fikrig E, Montgomery RR, Lord JM, Shaw AC. Human innate immunosenescence: causes and consequences for immunity in old age. Trends Immunol. 2009;30:325–333. doi: 10.1016/j.it.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng NP. Aging of the immune system: how much can the adaptive immune system adapt? Immunity. 2006;24:495–499. doi: 10.1016/j.immuni.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AC, Joshi S, Greenwood H, Panda A, Lord JM. Aging of the innate immune system. Curr Opin Immunol. 2010;22:507–513. doi: 10.1016/j.coi.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyugen J, Agrawal S, Gollapudi S, Gupta S. Impaired functions of peripheral blood monocyte subpopulations in aged humans. J Clin Immunol. 2010;30:806–813. doi: 10.1007/s10875-010-9448-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasto S, Candore G, Balistreri CR, Caruso M, Colonna-Romano G, Grimaldi MP, Listi F, Nuzzo D, Lio D, Caruso C. Inflammatory networks in ageing, age-related diseases and longevity. Mech Ageing Dev. 2007;128:83–91. doi: 10.1016/j.mad.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Bell JT, Tsai PC, Yang TP, Pidsley R, Nisbet J, Glass D, Mangino M, Zhai G, Zhang F, Valdes A. et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 2012;8:e1002629. doi: 10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garagnani P, Bacalini MG, Pirazzini C, Gori D, Giuliani C, Mari D, Di Blasio AM, Gentilini D, Vitale G, Collino S. et al. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell. 2012;11:1132–1134. doi: 10.1111/acel.12005. [DOI] [PubMed] [Google Scholar]
- Johansson A, Enroth S, Gyllensten U. Continuous aging of the human DNA Methylome throughout the human lifespan. PLoS One. 2013;8:e67378. doi: 10.1371/journal.pone.0067378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geretti E, Shimizu A, Kurschat P, Klagsbrun M. Site-directed mutagenesis in the B-neuropilin-2 domain selectively enhances its affinity to VEGF165, but not to semaphorin 3F. J Biol Chem. 2007;282:25698–25707. doi: 10.1074/jbc.M702942200. [DOI] [PubMed] [Google Scholar]
- Miao HQ, Lee P, Lin H, Soker S, Klagsbrun M. Neuropilin-1 expression by tumor cells promotes tumor angiogenesis and progression. Faseb J. 2000;14:2532–2539. doi: 10.1096/fj.00-0250com. [DOI] [PubMed] [Google Scholar]
- Bagri A, Tessier-Lavigne M, Watts RJ. Neuropilins in tumor biology. Clin Cancer Res. 2009;15:1860–1864. doi: 10.1158/1078-0432.CCR-08-0563. [DOI] [PubMed] [Google Scholar]
- Bielenberg DR, Pettaway CA, Takashima S, Klagsbrun M. Neuropilins in neoplasms: expression, regulation, and function. Exp Cell Res. 2006;312:584–593. doi: 10.1016/j.yexcr.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Bruder D, Probst-Kepper M, Westendorf AM, Geffers R, Beissert S, Loser K, von Boehmer H, Buer J, Hansen W. Neuropilin-1: a surface marker of regulatory T cells. Eur J Immunol. 2004;34:623–630. doi: 10.1002/eji.200324799. [DOI] [PubMed] [Google Scholar]
- Sarris M, Andersen KG, Randow F, Mayr L, Betz AG. Neuropilin-1 expression on regulatory T cells enhances their interactions with dendritic cells during antigen recognition. Immunity. 2008;28:402–413. doi: 10.1016/j.immuni.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozic G, Lupowitz Z, Zisapel N. Exonal elements and factors involved in the depolarization-induced alternative splicing of neurexin 2. J Mol Neurosci. 2012;50:221–233. doi: 10.1007/s12031-012-9919-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Siddiqui TJ, Huashan P, Yokomaku D, Hamdan FF, Champagne N, Lapointe M, Spiegelman D, Noreau A, Lafreniere RG. et al. Truncating mutations in NRXN2 and ia. Hum Genet. 2011;130:563–573. doi: 10.1007/s00439-011-0975-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou AS, Dooley J, Linterman MA, Pierson W, Ucar O, Kyewski B, Zuklys S, Hollander GA, Matthys P, Gray DH. et al. The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-alpha receptor. Nat Immunol. 2012;13:181–187. doi: 10.1038/ni.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- Santanam U, Zanesi N, Efanov A, Costinean S, Palamarchuk A, Hagan JP, Volinia S, Alder H, Rassenti L, Kipps T. et al. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proc Natl Acad Sci USA. 2010;107:12210–12215. doi: 10.1073/pnas.1007186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N. et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C. et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tserel L, Kolde R, Rebane A, Kisand K, Org T, Peterson H, Vilo J, Peterson P. Genome-wide promoter analysis of histone modifications in human monocyte-derived antigen presenting cells. BMC Genomics. 2010;11:642. doi: 10.1186/1471-2164-11-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The purification of monocyte cell population. The monocytes were analysed with FACSCalibur (BD Biosciences) using fluorescence conjugated antibodies against CD14 and CD3 (Miltenyi).