Abstract
Protein-DNA interactions are highly dependent upon salt such that the binding affinity precipitously decreases with increasing salt concentration in a phenomenon termed the polyelectrolyte effect. In this study, we provide evidence that the binding of EGR1 transcription factor to DNA displays virtually zero dependence on ionic strength under physiological salt concentrations and that such feat is accomplished via favorable enthalpic contributions. Importantly, we unearth the molecular origin of such favorable enthalpy and attribute it to the ability of H382 residue to stabilize the EGR1-DNA interaction via both intermolecular hydrogen bonding and van der Waals contacts against the backdrop of salt. Consistent with this notion, the substitution of H382 residue with other amino acids faithfully restores salt-dependent binding of EGR1 to DNA in a canonical fashion. Remarkably, H382 is highly conserved across other members of EGR family, implying that changes in bulk salt concentration are unlikely to play a significant role in modulating protein-DNA interactions central to this family of transcription factors. Taken together, our study reports the first example of a eukaryotic protein-DNA interaction capable of overriding the polyelectrolye effect.
Keywords: Protein-DNA thermodynamics, Salt tolerance, Hydrogen bonding, van der Waals contacts, Polyelectrolyte effect
INTRODUCTION
The charged groups in protein and DNA do not exist in isolation in water but are rather shielded with oppositely charged ions termed “counterions” so as to minimize electrostatic repulsions within their respective polarized regions. Upon association, the oppositely charged groups in protein and DNA engage in the formation of intermolecular salt bridges accompanied by the release of counterions into bulk water in a phenomenon that has come to be known as the polyelectrolyte effect (1–7). Given that the release of otherwise “immobilized” counterions into bulk water is concomitant with an increase in the degrees of freedom of the system, the polyelectrolyte effect generates a highly favorable entropic contribution to the free energy of binding of proteins to DNA. Because of their predominantly ionic nature, protein-DNA interactions are highly dependent upon salt such that the binding affinity precipitously decreases with increasing salt concentration (1–7). This is a hallmark of the polyelectrolyte effect in that increasing the concentration of bulk salt mitigates the liberation of counterions into bulk water.
While the polyelectrolyte effect is generally believed to be the major player involved in driving protein-DNA interactions, recent studies on bacterial DNA-binding proteins such as TBP and PolI challenge the generality of this school of thought (8, 9). On the contrary, increasing salt concentration augments rather than disrupts the binding of these bacterial proteins to DNA. Given the extreme environmental conditions that the bacteria have evolved to adopt, the reversal of such salt effect on protein-DNA interactions is perhaps not surprising. Nonetheless, the suggestion that the polyelectrolyte effect may not be a universal phenomenon in driving protein-DNA interactions and that certain protein-DNA interactions could actually thrive under high salt concentrations provokes further experimentation. Toward this goal, we set out here to uncover the nature of thermodynamic forces driving the binding of human EGR1 transcription factor, also known as Zif268, to its cognate DNA (10–12). Briefly, EGR1 is constructed on the classical TA-DB modular design, where the TA is the N-terminal transactivation domain and DB is the C-terminal DNA-binding domain (10, 12–14). Upon activation in response to extracellular stimuli—such as hormones, neurotransmitters and growth factors—EGR1 binds via its DB domain to the promoters of target genes containing the GCGTGGGCG consensus motif, referred to hereinafter as Zif268 response element (ZRE), in a sequence-dependent manner (15–17). The resulting EGR1-DNA interaction facilitates the TA domain to recruit a diverse array of transcriptional co-regulators to cognate DNA promoters and, in doing so, plays a key role in modulating the transcriptional machinery. Importantly, EGR1 couples extracellular stimuli to changes in gene expression responsible for a myriad of cellular activities ranging from cell growth and proliferation to apoptosis and oncogenic transformation (18–30).
In this study, we provide evidence that the binding of EGR1 transcription factor to DNA displays virtually zero dependence on ionic strength under physiological salt concentrations and that such feat is accomplished via favorable enthalpic contributions. Importantly, we unearth the molecular origin of such favorable enthalpy and attribute it to the ability of H382 residue to stabilize the EGR1-DNA interaction via both intermolecular hydrogen bonding and van der Waals contacts against the backdrop of salt. Consistent with this notion, the substitution of H382 residue with other amino acids faithfully restores salt-dependent binding of EGR1 to DNA in a canonical fashion. Remarkably, H382 is highly conserved across other members of EGR family, implying that changes in bulk salt concentration are unlikely to play a significant role in modulating protein-DNA interactions central to this family of transcription factors.
EXPERIMENTAL PROCEDURES
Protein preparation
The wildtype DB (DB_WT) domain (residues 331–430) of human EGR1 was cloned into pET30 bacterial expression vector with an N-terminal His-tag using Novagen LIC technology as described previously (31). The single-mutants of the DB domain of EGR1 containing H382A (DB_H382A), H382K (DB_H382K), H382R (DB_H382R) and H382E (DB_H382E) substitutions as well as the double-mutant containing the E354H/E410H (DB_HH) substitutions were generated through de novo DNA synthesis (GenScript Corporation) and subsequently cloned into pET30 bacterial expression as described for the DB_WT domain. All recombinant proteins were subsequently expressed in Escherichia coli BL21*(DE3) bacterial strain (Invitrogen) and purified on a Ni-NTA affinity column followed by size-exclusion chromatography (SEC) as described previously (31). It should be noted here that the BL21*(DE3) strain is optimized for high-yield expression of recombinant proteins in bacteria. Final yields were typically between 5–10mg protein of apparent homogeneity per liter of bacterial culture. Protein concentration was spectrophotometrically determined on the basis of extinction coefficients calculated using the online software ProtParam at ExPasy Server.
DNA synthesis
15-mer DNA oligos containing the ZRE consensus site (GCGTGGGCG) were commercially obtained from Sigma Genosys. The complete nucleotide sequence of the sense and antisense oligos constituting the ZRE duplex is shown below:
Oligo concentrations were determined spectrophotometrically on the basis of their extinction coefficients derived from their nucleotide sequences using the online software OligoAnalyzer 3.1. Equimolar amounts of sense and antisense oligos were mixed together and heated at 95°C for 10min and then allowed to cool to room temperature to obtain double-stranded DNA (dsDNA) annealed oligos (ZRE duplex).
ITC measurements
Isothermal titration calorimetry (ITC) experiments were performed on a TA Nano-ITC instrument. Briefly, wildtype and mutant DB domains of EGR1 and the ZRE duplex were dialyzed in 50mM Sodium phosphate and 5mM β-mercaptoethanol containing varying concentrations of Sodium chloride (NaCl), Potassium chloride (KCl) or Sodium acetate (NaAc) at pH 7.0. All experiments were initiated by injecting 25 × 10μl aliquots of 100–200μM of ZRE duplex from the syringe into the calorimetric cell containing 0.95ml of 10–20μM of DB domain solution at 25°C. The change in thermal power as a function of each injection was automatically recorded using the integrated NanoAnalyze software. The raw data were further integrated to yield binding isotherms of heat release per injection as a function of molar ratio of ZRE duplex to DB domains. The heats of mixing and dilution were subtracted from the heat of binding per injection by carrying out a control experiment in which the same buffer in the calorimetric cell was titrated against the ZRE duplex in an identical manner. To determine the equilibrium constant (Kd) and the enthalpy change (ΔH) associated with the binding of DB domains to DNA, the binding isotherms were iteratively fit to a built-in one-site model by non-linear least squares regression analysis using the integrated NanoAnalyze software as described previously (31, 32). The free energy change (ΔG) upon binding was calculated from the relationship:
| [1] |
where R is the universal molar gas constant (1.99 cal/mol/K) and T is the absolute temperature (298 K). The entropic contribution (TΔS) to the free energy of binding was calculated from the relationship:
| [2] |
where ΔH and ΔG are as defined above.
Molecular modeling
Molecular modeling (MM) was employed to build a structural model of the DB domain of EGR1 in complex with the 15-mer ZRE duplex using the corresponding crystal structure determined by Pavletich and Pabo (16) in the MODELLER software (33) as described earlier (31). A total of 100 structural models were calculated and the structure with the lowest energy, as judged by the MODELLER Objective Function, was selected for further analysis. The structural model was rendered using RIBBONS (34).
RESULTS and DISCUSSION
Binding of EGR1 to DNA largely tolerates the effect of salt
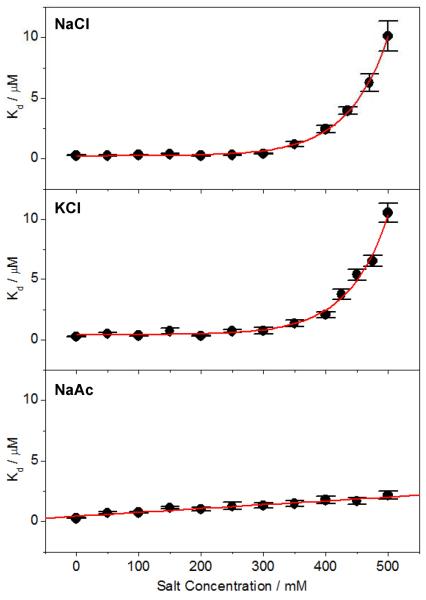
To investigate the effect of salt on EGR1-DNA interaction, we analyzed the binding of DB domain of EGR1 to a 15-mer ZRE duplex containing the GCGTGGGCG consensus motif as a function of various kosmotropic salts using ITC (Figure 1 and Tables 1–3). Our data reveal that the binding constant (Kd) is independent of NaCl concentration up to around 300mM and then drops off exponentially beyond this point (Figure 2 and Table 1). Remarkably, this behavior is also exquisitely reproduced in the presence of KCl (Figure 2 and Table 2), in which the sodium cation is replaced with potassium while keeping the chloride anion constant between these two salts. On the other hand, when the chloride anion is substituted with acetate while keeping the sodium cation constant, the lack of dependence of binding on salt is observed to be even more dramatic (Figure 2 and Table 3). Thus, while the binding of DB domain to DNA appears to be independent of NaCl and KCl salts up to about 300mM followed by an exponential decrease resulting in the reduction of binding by nearly 40-fold over the concentration range of 0–500mM, it displays a linear dependence on NaAc across the entire concentration range with an overall decrease in affinity of less than 10-fold. This salient observation strongly implies that the binding of DB domain of EGR1 to DNA not only displays much higher tolerance to NaAc compared to chloride salts but also partially restores its salt-dependence in a manner akin to classical protein-DNA interactions. How could that be so?
Figure 1.
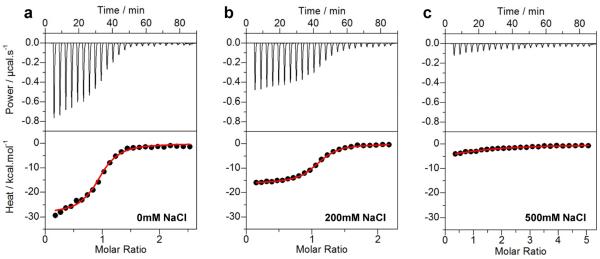
Representative ITC isotherms for the binding of ZRE duplex to the DB_WT domain of EGR1 at NaCl concentrations of 0mM (a), 200mM (b) and 500mM (c). The upper panels show raw ITC data expressed as change in thermal power with respect to time over the period of titration. In the lower panels, change in molar heat is expressed as a function of molar ratio of ZRE duplex to DB domain. The red solid lines in the lower panels show the fit of data to a one-site binding model using the integrated NanoAnalyze software as described previously (31, 32).
Table 1.
Thermodynamic parameters for the binding of wildtype DB domain (DB_WT) of EGR1 to DNA at pH 7 and 25°C under various NaCl concentrations
| [NaCl] / mM | Kd / nM | ΔH / kcal.mol−1 | TΔS / kcal.mol−1 | ΔG / kcal.mol−1 |
|---|---|---|---|---|
| 0 | 271 ± 35 | −26.70 ± 0.61 | −17.72 ± 0.69 | −8.97 ± 0.08 |
| 100 | 314 ± 39 | −19.01 ± 0.17 | −10.13 ± 0.24 | −8.88 ± 0.07 |
| 200 | 266 ± 66 | −16.32 ± 0.16 | −7.33 ± 0.31 | −8.99 ± 0.15 |
| 300 | 421 ± 49 | −14.27 ± 0.15 | −5.57 ± 0.22 | −8.71 ± 0.07 |
| 400 | 2441 ± 325 | −14.20 ± 0.72 | −6.54 ± 0.64 | −7.67 ± 0.08 |
| 500 | 10119 ± 1228 | −12.99 ± 0.37 | −6.17 ± 0.44 | −6.82 ± 0.07 |
The binding stoichiometries to the fits agreed to within ±10%. Errors were calculated from at least three independent measurements. All errors are given to one standard deviation.
Table 3.
Thermodynamic parameters for the binding of wildtype DB domain (DB_WT) of EGR1 to DNA at pH 7 and 25°C under various NaAc concentrations
| [NaAc] / mM | Kd / nM | ΔH / kcal.mol−1 | TΔS / kcal.mol−1 | ΔG / kcal.mol−1 |
|---|---|---|---|---|
| 0 | 271 ± 35 | −26.70 ± 0.61 | −17.72 ± 0.69 | −8.97 ± 0.08 |
| 100 | 755 ± 68 | −14.94 ± 0.69 | −6.58 ± 0.75 | −8.36 ± 0.06 |
| 200 | 1026 ± 132 | −12.99 ± 0.53 | −4.81 ± 0.61 | −8.18 ± 0.08 |
| 300 | 1330 ± 218 | −9.09 ± 0.21 | −1.06 ± 0.11 | −8.03 ± 0.10 |
| 400 | 1772 ± 308 | −6.58 ± 0.31 | +1.28 ± 0.41 | −7.86 ± 0.10 |
| 500 | 2174 ± 357 | −3.51 ± 0.19 | +4.22 ± 0.09 | −7.74 ± 0.10 |
The binding stoichiometries to the fits agreed to within ±10%. Errors were calculated from at least three independent measurements. All errors are given to one standard deviation.
Figure 2.
Dependence of equilibrium dissociation constant (Kd) on concentrations of NaCl (top panel), KCl (middle panel) and NaAc (bottom panel) for the binding of ZRE duplex to the DB_WT domain of EGR1. Note that the red solid lines represent exponential fits to data points in the top and middle panels, whilst the one in the bottom panel denotes linear regression fit. The error bars were calculated from at least three independent measurements to one standard deviation.
Table 2.
Thermodynamic parameters for the binding of wildtype DB domain (DB_WT) of EGR1 to DNA at pH 7 and 25°C under various KCl concentrations
| [KCl] / mM | Kd / nM | ΔH / kcal.mol−1 | TΔS / kcal.mol−1 | ΔG / kcal.mol−1 |
|---|---|---|---|---|
| 0 | 271 ± 35 | −26.70 ± 0.61 | −17.72 ± 0.69 | −8.97 ± 0.08 |
| 100 | 345 ± 47 | −17.31 ± 0.75 | −8.48 ± 0.67 | −8.83 ± 0.08 |
| 200 | 324 ± 50 | −11.33 ± 0.25 | −2.47 ± 0.15 | −8.87 ± 0.09 |
| 300 | 758 ± 265 | −6.78 ± 0.26 | +1.60 ± 0.47 | −8.37 ± 0.21 |
| 400 | 2070 ± 251 | −5.95 ± 0.55 | +1.81 ± 0.48 | −7.76 ± 0.07 |
| 500 | 10534 ± 784 | −4.45 ± 0.23 | +2.35 ± 0.18 | −6.80 ± 0.04 |
The binding stoichiometries to the fits agreed to within ±10%. Errors were calculated from at least three independent measurements. All errors are given to one standard deviation.
According to the Hofmeister series (35–37), acetate harbors a relatively higher degree of kosmotropic character than chloride. In the context of protein structure and stability, this implies that acetate bears higher potential than chloride to drive intermolecular interactions by virtue of its ability to cause water molecules to interact with apolar surfaces on proteins more favorably so as to become more “ordered”. Upon binding to DNA, the release of such “ordered” water molecules from apolar surfaces would be highly entropically favorable such that it could suppress the effect of increasing salt concentration in the bulk water. The greater the kosmotropic nature of a salt the greater suppressing power it is likely to exert. Accordingly, the ability of EGR1-DNA interaction to display higher tolerance to NaAc could be ascribed to its higher kosmotropic character relative to NaCl and KCl. In short, our data presented above strongly argue that the binding of EGR1 to DNA is independent of physiological concentrations of salt. This finding is thus in a marked contrast to the more ubiquitous polyelectrolyte effect that is in general believed to drive protein-DNA interactions (1–7). In this classical model, the affinity of protein-DNA interactions rapidly drops with increasing salt concentration. Notably, such reduction in binding affinity is attributed to the decrease in favorable entropy of counterion release from DNA as the bulk concentration of salt is increased (7, 38, 39). In the case of EGR1-DNA interaction, such loss of entropy due to counterion release under physiological concentrations of salt would be compensated either by an equal gain of favorable entropy due to the release of ordered water from apolar surfaces into bulk phase and/or a compensatory gain of favorable enthalpy due to the formation of non-ionic interactions such as hydrogen bonding and van der Waals contacts.
Favorable enthalpic factors account for the salt tolerance of the binding of EGR1 to DNA
To understand how an interplay between underlying thermodynamic forces confers upon EGR1-DNA interaction the ability to override the polyelectrolyte effect, we next analyzed the dependence of enthalpic (ΔH) and entropic (TΔS) contributions to the free energy (ΔG) of binding as a function of salt concentration (Figure 3). Our analysis shows that increasing salt concentration strongly correlates with an increase in the entropic contribution to the free energy of binding of EGR1 to DNA (Figures 3a–3c). In sharp contrast, this favorable entropic contribution is exquisitely mirrored by an equal but an unfavorable decrease in enthalpic contribution as a function of concentration of all three salts. Thus, the overall effect of such ΔH-TΔS compensation results in little or no change in ΔG up to about 300mM of chloride salts. Remarkably, after this point, the unfavorable enthalpic contributions begin to considerably outweigh the favorable entropic contributions such that this breakdown in ΔH-TΔS compensation after about 300mM is directly reflected in the loss of free energy of EGR1-DNA interaction. A similar but perhaps a somewhat more subtle trend is also observed in the case of increasing concentration of NaAc.
Figure 3.
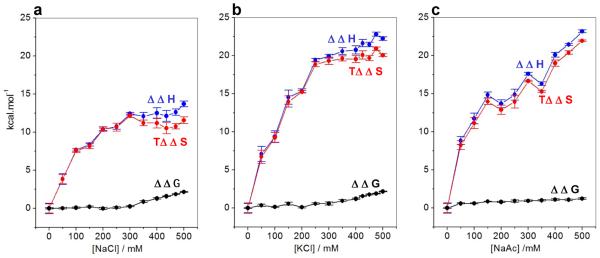
Effect of increasing concentrations of NaCl (a), KCl (b) and NaAc (c) on the change in the contributions of enthalpic (ΔΔH) ( ) and entropic (TΔΔS) (
) and entropic (TΔΔS) ( ) forces to the free energy (ΔΔG) (●) with respect to the corresponding contributions in the absence of salt accompanying the binding of ZRE duplex to DB_WT domain of EGR1. Note that an increasing value of ΔΔH is indicative of a decrease in the favorable enthalpic contribution, an increasing value of TΔΔS is indicative of an increase in the favorable entropic contribution, and an increasing value of ΔΔG is indicative of a decrease in the overall free energy of binding. The solid lines are merely used to connect data points for clarity. The error bars were calculated from at least three independent measurements to one standard deviation.
) forces to the free energy (ΔΔG) (●) with respect to the corresponding contributions in the absence of salt accompanying the binding of ZRE duplex to DB_WT domain of EGR1. Note that an increasing value of ΔΔH is indicative of a decrease in the favorable enthalpic contribution, an increasing value of TΔΔS is indicative of an increase in the favorable entropic contribution, and an increasing value of ΔΔG is indicative of a decrease in the overall free energy of binding. The solid lines are merely used to connect data points for clarity. The error bars were calculated from at least three independent measurements to one standard deviation.
In sum, the decrease in the binding affinity of EGR1 to DNA under high concentrations of salt is overall due to the loss of favorable enthalpic contributions as opposed to entropic. In retrospect, this strongly implies that the thermodynamic basis of the ability of EGR1-DNA interaction to resist the effect of physiological concentrations of salt must reside in underlying enthalpic factors such as hydrogen bonding and van der Waals contacts in lieu of entropic forces.
Physical basis of the salt independence of EGR1-DNA interaction lies in the formation of intermolecular hydrogen bonding and van der Waals contacts
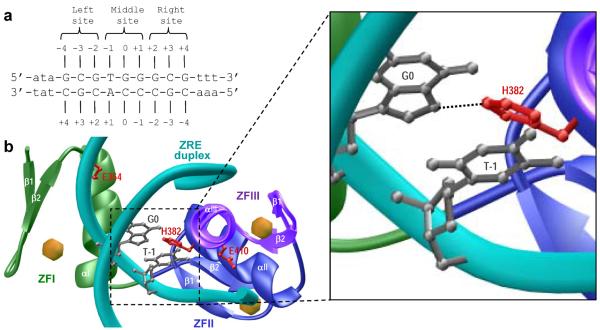
In order to elucidate the physical basis of the favorable enthalpy driving the binding of EGR1 to DNA, we next generated a structural model of the DB domain in complex with the ZRE duplex on the basis of known crystal structure determined by Pavletich and Pabo (16). As shown in Figure 4, the EGR1-DNA interaction is driven by the binding of DB domain as a monomer to the major groove within the ZRE duplex. Notably, the DB domain of EGR1 is comprised of three tandem copies of C2H2-type zinc fingers (40–42), designated herein ZFI, ZFII and ZFIII, which come together in space to assemble into an arc-like architecture that snugly fits into the major groove of DNA. Each zinc finger within the DB domain is comprised of an α-helix and an antiparallel double-stranded β-sheet (β1–β2) that together sandwich a Zn2+ divalent ion, the latter being coordinated in a tetrahedral arrangement by two histidine residues and two cysteine residues. It is telling that the EGR1-DNA interaction is driven by the binding of each zinc finger to one of the three subsites, each subsite being comprised of a trinucleotide sequence, within the 9-bp GCGTGGGCG consensus motif (Figures 4a and 4b). In particular, at each of the three subsites within the ZRE duplex occupied by one of the three zinc fingers, the protein-DNA contacts are largely afforded by the α-helix, which fits into the major groove of DNA, and β2-strand, which contacts the DNA phosphate backbone. On the other hand, the β1-strand appears to serve as a scaffold and does not directly engage in intermolecular contacts with DNA.
Figure 4.
Structural analysis of EGR1-DNA interactions. (a) ZRE duplex containing the GCGTGGGCG tripartite consensus motif. The consensus nucleotides within this motif are capitalized whilst the flanking nucleotides are shown in small letters. The three sub-sites within the consensus motif are marked for clarity and accommodate ZFI (right site), ZFII (middle site) and ZFIII (left site) within the DB domain. The numbering of various nucleotides with respect to the central nucleotide of the middle site (which is arbitrarily assigned zero) are indicated. (b) Structural model of the DB domain of EGR1 in complex with ZRE duplex containing the GCGTGGGCG consensus sequence. Note that the DB domain is comprised of three tandem C2H2-type zinc fingers, designated herein ZFI (green), ZFII (blue) and ZFIII (magenta). Each zinc finger is comprised of an α-helix and a double-stranded antiparallel β-sheet (β1-β2) that together sandwich a Zn2+ divalent ion indicated by a sphere (yellow). The DNA backbone is shown in cyan and the bases are omitted for clarity, except for G0 and T-1 (shown in gray) within the sense strand that engage in intermolecular contacts with the imidazole ring of H382 (colored red) located within the α-helix (αII) of ZFII. The expanded window offers a close-up view of these key intermolecular contacts and the putative hydrogen bond between the Hο2 atom of H382 and N7 atom of G0 is indicated by a dashed line. The sidechain moieties of amino acid residues E354 (ZFI) and E410 (ZFIII), the structural-equivalents of H382 (ZFII) within the flanking zinc fingers of the DB domain, are also shown and colored red for comparison.
Importantly, the binding of DB domain of EGR1 appears to be strongly governed by numerous van der Waals contacts in addition to an extensive network of intermolecular hydrogen bonding and ion pairing (16). In particular, detailed examination of the atomic structure of the DB domain of EGR1 in complex with the ZRE duplex shows that an histidine residue (H382), located within the first turn of α-helix (αII) of ZFII but not involved in coordinating the zinc ligand, protrudes deep into the major groove at the protein-DNA interface (Figure 4b). Most significantly, the imidazole ring of H382 is coplanar with G0, the central guanine of the middle trinucleotide subsite that accommodates ZFII within the DB domain, and stacks against the pyrimidine ring of T-1 within the ZRE duplex. In this manner, H382 plays a key role in stabilizing the EGR1-DNA interaction via a two-prong mechanism. Firstly, the coplanar alignment of the imidazole ring of H382 with the purine ring of G0 facilitates the formation of an hydrogen bond between the Hε2 atom of H382 and N7 atom of G0. Secondly, stacking of the imidazole ring of H382 against the pyrimidine ring of T-1 promotes van der Waals contacts between the protein and DNA. In light of these observations, the ability of NaAc to partially restore salt-dependent binding of EGR1 to DNA under physiological concentrations could be explained in two ways (Figure 2): (i) the carbonyl moiety of acetate may compete with the imidazole ring of H382 for hydrogen bonding with G0; and (ii) the methyl group of acetate may compete with the imidazole ring of H382 for van der Waals contacts with T-1. Accordingly, the polar and apolar characters of acetate anion are likely to disrupt the hydrogen bonding and stacking interactions of H382 with DNA.
Notably, H382 located within ZFII is replaced by a glutamate residue in ZFI (E354) and ZFIII (E410) at the structurally-equivalent positions. Thus, the lack of conservation of H382 in ZFI and ZFIII implicates a unique role of ZFII in dictating the binding of EGR1 to DNA. In our earlier work (31), we indeed demonstrated that the binding of EGR1 to DNA is tightly modulated by solution pH by virtue of the ability of the imidazole ring of H382 to undergo protonation-deprotonation-coupled equilibrium. Could the ability of H382 to engage in key intermolecular hydrogen bonding and van der Waals contacts with DNA also account for the favorable enthalpy that could ultimately confer upon EGR1-DNA interaction the ability to tolerate physiological concentrations of salt?
Salt tolerance of EGR1-DNA interaction is solely attributable to H382
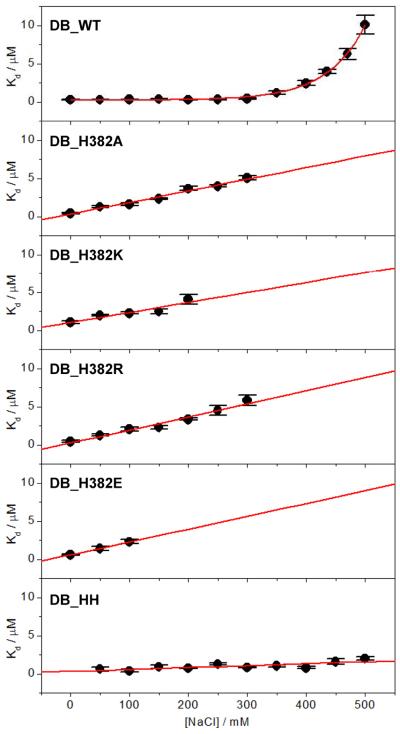
To test our hypothesis that the ability of EGR1 to bind to DNA in a salt-independent manner is attributable to H382 by virtue of its ability to engage in key intermolecular hydrogen bonding and van der Waals contacts, we introduced and measured the effect of the binding of DB domain containing the H382A substitution (DB_H382A) to DNA. We reasoned that the H382A substitution should be expected to remove the contribution of the imidazole ring to the free energy of binding and thereby eliminate the salt tolerance of the binding of EGR1 to DNA. Consistent with our rationale, our analysis reveals that while increasing NaCl concentration has little or negligible effect on the binding of wildtype DB (DB_WT) domain to DNA, the binding of DB_H382A domain displays a sharp linear dependence such that the affinity drops by more than an order of magnitude over the concentration window of 0–300mM in a manner akin to classical protein-DNA interactions under the control of polyelectrolyte effect (Figure 5). To further corroborate our hypothesis, we also introduced and measured the effect of the binding of DB domain of EGR1 containing the H382K (DB_H382K) and H382R (DB_H382R) substitutions to DNA. In light of our previous study showing that the protonation-deprotonation of H382 tightly modulates the binding of EGR1 to DNA (31), these basic substitutions were introduced to mimic the effect of a protonated histidine containing a net positive charge at H382. As expected, the binding of both DB_H382K and DB_H382R domains to DNA exhibited strong NaCl-dependence in a manner similar to the DB_H382A construct over the concentration range of 0–300mM (Figure 5).
Figure 5.
Dependence of Kd on increasing NaCl concentration for the binding of ZRE duplex to DB_WT, DB_H382A, DB_H382K, DB_H382R, DB_H382E and DB_HH domains of EGR1. Note that the red solid line represents an exponential fit to data points in the top panel, whilst it denotes linear regression fits in other panels. The error bars were calculated from at least three independent measurements to one standard deviation.
Given that the H382 residue located within the ZFII of DB domain of EGR1 is replaced by a glutamate residue at the structurally-equivalent positions within ZFI (E354) and ZFIII (E410) (Figure 4), we also wondered how H382E substitution might influence the binding of DB domain to DNA. Toward this goal, we introduced and measured the effect of the binding of DB domain of EGR1 containing the H382E substitution (DB_H382E) to DNA. Consistent with our hypothesis that H382 compensates the effect of salt on the binding of DB domain to DNA, our data reveal that the binding of DB_H382E domain to DNA is highly dependent upon NaCl within the concentration window of 0–100mM though measurements at higher salt concentrations were not possible (Figure 5). We note that while analysis of the binding of various mutant DB domains to DNA was only feasible up to NaCl concentrations of about 300mM due to low enthalpic change associated with binding under high salt concentrations, our data nonetheless are strongly suggestive that H382 solely accounts for the compensatory effect of salt on EGR1-DNA interaction. Indeed, if this hypothesis is valid, we would expect the E354H and E410H substitutions to augment the compensatory effect of salt on the binding of DB domain of EGR1 to DNA. To accomplish this goal, we introduced and measured the effect of the binding of DB domain of EGR1 containing the E354H/E410H double-substitution (DB_HH) to DNA. Interestingly, while salt has little or no effect on the binding of DB_WT domain to DNA only up to about 300mM of NaCl, the DB_HH domain binds to DNA flawlessly with little or no change in affinity even at concentrations of 500mM (Figure 5).
It is also noteworthy that the introduction of various point mutations in place of H382 within the DB domain is concomitant with the loss of binding affinity by several folds (Table 4), implying that these mutations perturb protein structure to a varying degree. However, addition of salt further reduces this affinity in a manner akin to that observed for classical protein-DNA interactions (Figure 5). The detrimental effect of aforementioned mutations on binding thus further implicates a critical role of H382 in mediating EGR1-DNA interactions. Surprisingly, while the E354H and E410H substitutions augment the compensatory effect of salt on the binding of DB domain to DNA (Figure 5), they are met with a slight loss of binding affinity (Table 4). This strongly argues that H382 plays a unique and non-equivalent role within the ZFII of DB domain with respect to its structural-equivalents E354 and E410 located within ZFI and ZFIII, respectively. Taken together, our analysis of the effect of various mutations on the binding of DB domain bears important consequences on the rationale design of zinc fingers for therapeutic intervention.
Table 4.
Thermodynamic parameters for the binding of wildtype (DB_WT) and various mutant DB domains of EGR1 to DNA at pH 7 and 25°C
| Variants | Kd / nM | ΔH / kcal.mol−1 | TΔS / kcal.mol−1 | ΔG / kcal.mol−1 |
|---|---|---|---|---|
| DB_WT | 314 ± 39 | −19.01 ± 0.17 | −10.13 ± 0.24 | −8.88 ± 0.07 |
| DB_H382A | 1614 ± 190 | −9.42 ± 0.47 | −1.51 ± 0.40 | −7.91 ± 0.07 |
| DB_H382K | 2257 ± 133 | −4.59 ± 0.14 | 3.12 ± 0.17 | −7.71 ± 0.04 |
| DB_H382R | 2041 ± 253 | −6.52 ± 0.47 | 1.25 ± 0.40 | −7.77 ± 0.07 |
| DB_H382E | 2298 ± 270 | −1.60 ± 0.20 | 5.97 ± 0.27 | −7.70 ± 0.07 |
| DB_HH | 530 ± 69 | −7.26 ± 0.55 | 1.31 ± 0.62 | −8.57 ± 0.08 |
All measurements were conducted in 50mM Sodium phosphate and 5mM β-mercaptoethanol containing 100mM NaCl. The binding stoichiometries to the fits agreed to within ±10%. Errors were calculated from at least three independent measurements. All errors are given to one standard deviation.
Loss of enthalpy is largely responsible for the loss of salt tolerance of the binding of EGR1 to DNA
Our data presented above strongly suggest that the molecular origin of the ability of EGR1 to tolerate physiological concentrations of salt upon binding to DNA solely resides in the ability of H382 to engage in key intermolecular hydrogen bonding and van der Waals contacts. Consistent with this argument, the linear decrease in the affinity of binding of DB_H382A mutant domain to DNA on NaCl concentration in the 0–300mM range appears to largely result from the loss of enthalpic factors relative to DB_WT domain (Figure 6). Thus, while increasing NaCl concentration results in the loss of enthalpic contribution for both DB_WT and DB_H382A domains, such loss is greater for the latter (Figure 6a). On the other hand, the entropic gain with increasing NaCl concentration is only marginally larger for the binding of DB_H382A domain relative to DB_WT (Figure 6b). Thus, the loss in the free energy of binding of DB_H382A domain to DNA with increasing NaCl concentration can be largely attributed to the loss of favorable enthalpic contributions (Figure 6c), while DB_WT domain appears to tolerate the effect of NaCl over the same concentration window. Importantly, the decrease in the binding affinity of DB_H382K, DB_H382R and DB_H382E mutant domains to DNA is also attributable to the greater loss of enthalpic contributions to the free energy relative to DB_WT. Conversely, the binding of DB_HH domain to DNA with little or no change in affinity even at concentrations of 500mM NaCl is concomitant with significantly much less loss of favorable enthalpy relative to DB_WT domain. Taken together, these observations are clearly indicative of the fact that the removal of imidazole moiety of H382 most likely leads to disruption of intermolecular hydrogen bonding and van der Waals contacts between EGR1 and DNA and that such disruption culminates in the greater loss of enthalpy.
Figure 6.
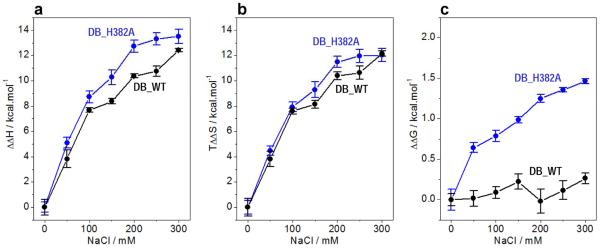
Comparison of enthalpic (ΔΔH) (a) and entropic (TΔΔS) (b) contributions to the free energy (ΔΔG) (c) with respect to the corresponding contributions in the absence of salt accompanying the binding of ZRE duplex to DB_WT (●) and DB_H382 ( ) domains of EGR1 as a function of increasing NaCl concentration. Note that an increasing value of ΔΔH is indicative of a decrease in the favorable enthalpic contribution, an increasing value of TΔΔS is indicative of an increase in the favorable entropic contribution, and an increasing value of ΔΔG is indicative of a decrease in the overall free energy of binding. The solid lines are merely used to connect data points for clarity. The error bars were calculated from at least three independent measurements to one standard deviation.
) domains of EGR1 as a function of increasing NaCl concentration. Note that an increasing value of ΔΔH is indicative of a decrease in the favorable enthalpic contribution, an increasing value of TΔΔS is indicative of an increase in the favorable entropic contribution, and an increasing value of ΔΔG is indicative of a decrease in the overall free energy of binding. The solid lines are merely used to connect data points for clarity. The error bars were calculated from at least three independent measurements to one standard deviation.
CONCLUSIONS
The conventional wisdom in macromolecular biophysics has it that the polyelectrolyte effect is the major player involved in driving protein-DNA interactions (1–7). It would indeed be preposterous to suggest otherwise. Of thousands of biophysical studies conducted on protein-DNA interactions over the past couple of decades or so, only two cases have been hitherto documented where increasing salt concentration did not correlate with the disruption of this key intermolecular association (8, 9). Our study thus reported here not only adds to the growing notion that the polyelectrolyte effect is not a universal phenomenon in driving protein-DNA interactions but should also open up further dialogue and scientific investigation into the molecular basis of protein-DNA recognition. What is so remarkable about EGR1-DNA interaction is that the DB domain bears the hallmarks of a classical DNA-binding protein in that it is electrostatically polarized such that the face of the molecule that snuggles into major groove of DNA is overall positively charged while the opposite face predominantly harbors a neutral surface (16, 31). Yet, against this backdrop of a potentially overwhelming electrostatic force, the EGR1-DNA interaction appears to resist the effect of physiological concentrations of salt. Most significantly, we have demonstrated here that the molecular origin of such salt tolerance lies in the release of favorable enthalpy due solely to the ability of H382—located within a sea of basic residues—to engage in key intermolecular hydrogen bonding and van der Waals contacts.
We previously reported that while the H382 residue within ZFII of EGR1 is fully conserved in all other EGR members (EGR2–EGR4), it is replaced by a glutamate at the structurally-equivalent position within ZFII of all members (KLF1–KLF17) of the closely-related KLF family (31). On the other hand, while the glutamate residues located at the structurally-equivalent position to H382 within ZFI (E354) and ZFIII (E410) of EGR1 are fully conserved within other members of EGR family, they are replaced by an histidine residue within all KLF members. Given the importance of H382 in driving the binding of EGR1 to DNA in a salt-independent manner, it is tempting to speculate that the binding of all other members of EGR and KLF families to DNA must also occur via a similar mechanism and that these related protein-DNA interactions must also bear the potential to suppress the effect of salt under physiological concentrations. It is also conceivable that the KLF members may have evolved to resist the effect of salt on their ability to bind to DNA even more strongly than the members of EGR family. This notion is supported by the observation that the binding of the DB_HH domain of EGR1 to DNA shows much higher tolerance to salt concentrations than the DB_WT domain. Interestingly, our study bears important implications on the physiological context of EGR-DNA and KLF-DNA interactions within the nucleus of the living cell, where slight changes in bulk salt should be expected to have profound consequences on the ability of transcription factors to operate. In fact, bulk salt is likely to serve as an allosteric switch to regulate protein-DNA interactions within the cell nucleus. Accordingly, the apparent ability of EGR and KLF members to by-pass this regulatory mechanism likely suggests a key role of these transcription factors in divergent cellular processes.
In conclusion, our study offers rare insights into the ability of a transcription factor to tolerate physiological concentrations of salt in binding to its cognate DNA. Given that such phenomenon has only been previously demonstrated for bacterial DNA-binding proteins (8, 9), our study is even more remarkable in that it represents the first example of a eukaryotic protein-DNA interaction capable of overriding the polyelectrolye effect.
ACKNOWLEDGMENTS
This work was supported by funds from the National Institutes of Health (Grant# R01-GM083897) and the USylvester Braman Family Breast Cancer Institute to AF. CBM is a recipient of a postdoctoral fellowship from the National Institutes of Health (Award# T32-CA119929).
ABBREVIATIONS
- DB
DNA-binding (domain)
- EGR
Early growth response
- EGR1
Early growth response (protein) 1
- ITC
Isothermal titration calorimetry
- KLF
Krueppel-like factor
- LIC
Ligation-independent cloning
- MM
Molecular modeling
- PolI
DNA polymerase I
- TA
Transactivation (domain)
- TBP
TATA-box binding protein
- ZF
Zinc finger
- ZRE
Zif268 (EGR1) response element
REFERENCES
- 1.Record MT, Jr., Lohman ML, De Haseth P. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976;107:145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- 2.Record MT, Jr., Anderson CF, Lohman TM. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q Rev Biophys. 1978;11:103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- 3.Record MT, Jr., Anderson CF, Mills P, Mossing M, Roe JH. Ions as regulators of protein-nucleic acid interactions in vitro and in vivo. Adv Biophys. 1985;20:109–135. doi: 10.1016/0065-227x(85)90033-4. [DOI] [PubMed] [Google Scholar]
- 4.Lohman TM, Mascotti DP. Thermodynamics of ligand-nucleic acid interactions. Methods Enzymol. 1992;212:400–424. doi: 10.1016/0076-6879(92)12026-m. [DOI] [PubMed] [Google Scholar]
- 5.Record MT, Jr., Ha JH, Fisher MA. Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability and specificity and mechanism of formation of site-specific complexes between proteins and helical DNA. Methods Enzymol. 1991;208:291–343. doi: 10.1016/0076-6879(91)08018-d. [DOI] [PubMed] [Google Scholar]
- 6.Manning GS. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 7.Sharp KA, Friedman RA, Misra V, Hecht J, Honig B. Salt effects on polyelectrolyte-ligand binding: comparison of Poisson-Boltzmann, and limiting law/counterion binding models. Biopolymers. 1995;36:245–262. doi: 10.1002/bip.360360211. [DOI] [PubMed] [Google Scholar]
- 8.O'Brien R, DeDecker B, Fleming KG, Sigler PB, Ladbury JE. The effects of salt on the TATA binding protein-DNA interaction from a hyperthermophilic archaeon. J Mol Biol. 1998;279:117–125. doi: 10.1006/jmbi.1998.1743. [DOI] [PubMed] [Google Scholar]
- 9.Deredge DJ, Baker JT, Datta K, Licata VJ. The glutamate effect on DNA binding by pol I DNA polymerases: osmotic stress and the effective reversal of salt linkage. J Mol Biol. 2010;401:223–238. doi: 10.1016/j.jmb.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Gashler A, Sukhatme VP. Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Prog Nucleic Acid Res Mol Biol. 1995;50:191–224. doi: 10.1016/s0079-6603(08)60815-6. [DOI] [PubMed] [Google Scholar]
- 11.Iwaki K, Sukhatme VP, Shubeita HE, Chien KR. Alpha- and beta- adrenergic stimulation induces distinct patterns of immediate early gene expression in neonatal rat myocardial cells. fos/jun expression is associated with sarcomere assembly; Egr-1 induction is primarily an alpha 1-mediated response. J Biol Chem. 1990;265:13809–13817. [PubMed] [Google Scholar]
- 12.Sukhatme VP. Early transcriptional events in cell growth: the Egr family. J Am Soc Nephrol. 1990;1:859–866. doi: 10.1681/ASN.V16859. [DOI] [PubMed] [Google Scholar]
- 13.Christy BA, Lau LF, Nathans D. A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with “zinc finger” sequences. Proc Natl Acad Sci U S A. 1988;85:7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pabo CO, Sauer RT. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 15.Christy B, Nathans D. DNA binding site of the growth factor-inducible protein Zif268. Proc Natl Acad Sci U S A. 1989;86:8737–8741. doi: 10.1073/pnas.86.22.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton TB, Borel F, Romaniuk PJ. Comparison of the DNA binding characteristics of the related zinc finger proteins WT1 and EGR1. Biochemistry. 1998;37:2051–2058. doi: 10.1021/bi9717993. [DOI] [PubMed] [Google Scholar]
- 18.Yan SF, Fujita T, Lu J, Okada K, Shan Zou Y, Mackman N, Pinsky DJ, Stern DM. Egr-1, a master switch coordinating upregulation of divergent gene families underlying ischemic stress. Nat Med. 2000;6:1355–1361. doi: 10.1038/82168. [DOI] [PubMed] [Google Scholar]
- 19.Thiel G, Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol. 2002;193:287–292. doi: 10.1002/jcp.10178. [DOI] [PubMed] [Google Scholar]
- 20.Ahmed MM. Regulation of radiation-induced apoptosis by early growth response-1 gene in solid tumors. Curr Cancer Drug Targets. 2004;4:43–52. doi: 10.2174/1568009043481704. [DOI] [PubMed] [Google Scholar]
- 21.Adamson ED, Mercola D. Egr1 transcription factor: multiple roles in prostate tumor cell growth and survival. Tumour Biol. 2002;23:93–102. doi: 10.1159/000059711. [DOI] [PubMed] [Google Scholar]
- 22.Gregg J, Fraizer G. Transcriptional Regulation of EGR1 by EGF and the ERK Signaling Pathway in Prostate Cancer Cells. Genes Cancer. 2011;2:900–909. doi: 10.1177/1947601911431885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Virolle T, Krones-Herzig A, Baron V, De Gregorio G, Adamson ED, Mercola D. Egr1 promotes growth and survival of prostate cancer cells. Identification of novel Egr1 target genes. J Biol Chem. 2003;278:11802–11810. doi: 10.1074/jbc.M210279200. [DOI] [PubMed] [Google Scholar]
- 24.Virolle T, Adamson ED, Baron V, Birle D, Mercola D, Mustelin T, de Belle I. The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat Cell Biol. 2001;3:1124–1128. doi: 10.1038/ncb1201-1124. [DOI] [PubMed] [Google Scholar]
- 25.Krones-Herzig A, Mittal S, Yule K, Liang H, English C, Urcis R, Soni T, Adamson ED, Mercola D. Early growth response 1 acts as a tumor suppressor in vivo and in vitro via regulation of p53. Cancer Res. 2005;65:5133–5143. doi: 10.1158/0008-5472.CAN-04-3742. [DOI] [PubMed] [Google Scholar]
- 26.Huang RP, Fan Y, de Belle I, Niemeyer C, Gottardis MM, Mercola D, Adamson ED. Decreased Egr-1 expression in human, mouse and rat mammary cells and tissues correlates with tumor formation. Int J Cancer. 1997;72:102–109. doi: 10.1002/(sici)1097-0215(19970703)72:1<102::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 27.Joslin JM, Fernald AA, Tennant TR, Davis EM, Kogan SC, Anastasi J, Crispino JD, Le Beau MM. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007;110:719–726. doi: 10.1182/blood-2007-01-068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zagurovskaya M, Shareef MM, Das A, Reeves A, Gupta S, Sudol M, Bedford MT, Prichard J, Mohiuddin M, Ahmed MM. EGR-1 forms a complex with YAP-1 and upregulates Bax expression in irradiated prostate carcinoma cells. Oncogene. 2009;28:1121–1131. doi: 10.1038/onc.2008.461. [DOI] [PubMed] [Google Scholar]
- 29.Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006;13:115–124. doi: 10.1038/sj.cgt.7700896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calogero A, Arcella A, De Gregorio G, Porcellini A, Mercola D, Liu C, Lombari V, Zani M, Giannini G, Gagliardi FM, Caruso R, Gulino A, Frati L, Ragona G. The early growth response gene EGR-1 behaves as a suppressor gene that is down-regulated independent of ARF/Mdm2 but not p53 alterations in fresh human gliomas. Clin Cancer Res. 2001;7:2788–2796. [PubMed] [Google Scholar]
- 31.Mikles DC, Bhat V, Schuchardt BJ, Deegan BJ, Seldeen KL, McDonald CB, Farooq A. pH modulates the binding of early growth response protein 1 transcription factor to DNA. FEBS J. 2013;280:3669–3684. doi: 10.1111/febs.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 33.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative Protein Structure Modeling of Genes and Genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 34.Carson M. Ribbons 2.0. J. Appl. Crystallogr. 1991;24:958–961. [Google Scholar]
- 35.Baldwin RL. How Hofmeister ion interactions affect protein stability. Biophys J. 1996;71:2056–2063. doi: 10.1016/S0006-3495(96)79404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cacace MG, Landau EM, Ramsden JJ. The Hofmeister series: salt and solvent effects on interfacial phenomena. Q Rev Biophys. 1997;30:241–277. doi: 10.1017/s0033583597003363. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Cremer PS. Interactions between macromolecules and ions: The Hofmeister series. Curr Opin Chem Biol. 2006;10:658–663. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 38.Ha JH, Capp MW, Hohenwalter MD, Baskerville M, Record MT., Jr. Thermodynamic stoichiometries of participation of water, cations and anions in specific and non-specific binding of lac repressor to DNA. Possible thermodynamic origins of the “glutamate effect” on protein-DNA interactions. J Mol Biol. 1992;228:252–264. doi: 10.1016/0022-2836(92)90504-d. [DOI] [PubMed] [Google Scholar]
- 39.Lohman TM, Overman LB, Ferrari ME, Kozlov AG. A highly salt-dependent enthalpy change for Escherichia coli SSB protein-nucleic acid binding due to ion-protein interactions. Biochemistry. 1996;35:5272–5279. doi: 10.1021/bi9527606. [DOI] [PubMed] [Google Scholar]
- 40.Green A, Parker M, Conte D, Sarkar B. Zinc Finger Proteins: A Bridge Between Transition Metals and Gene Regulation. J Trace Element Exp Med. 1998;11:103–118. [Google Scholar]
- 41.Green A, Sarkar B. Alteration of zif268 zinc-finger motifs gives rise to non-native zinc-co-ordination sites but preserves wild-type DNA recognition. Biochem J. 1998;333(Pt 1):85–90. doi: 10.1042/bj3330085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]