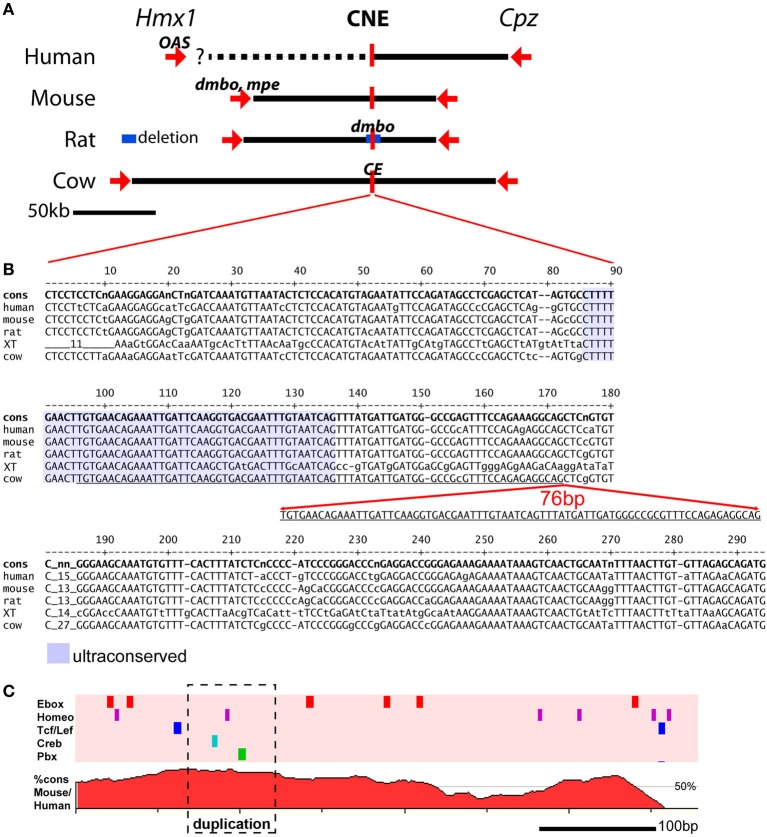
Figure 2.
Genomic location of mutations affecting pinna development at the Hmx1 locus. (A) The chromosomal relationship between the Hmx1 gene (and in some species not shown, two related Hmx genes), a distal Hmx1 CNE, and the neighboring gene encoding carboxypeptidase Z (Cpz) is conserved in all known vertebrate genomes (Quina et al., 2012b). The human OAS allele is a 26 bp deletion in the coding region of Hmx1, resulting in a frameshift, which is likely to be a null allele (Schorderet et al., 2008). The mouse dmbo and mpe alleles also map to the Hmx1 coding region, and are also likely to be null alleles (Jiang et al., 2002; Munroe et al., 2009). In contrast, the rat dmbo allele is a 5777 bp deletion encompassing the CNE (Chieffo et al., 1997), and the bovine crop ear (CE) allele is a 76 bp duplication within the CNE (Koch et al., 2013). (B) Alignment of the Hmx1 distal CNE sequence from the human, mouse, rat, xenopus tropicalis and cow genomes. Conserved bases are shown in upper case. Alignment of the x. tropicalis sequence with the mammalian genomes identifies a core “ultraconserved” sequence (shaded area) that can be identified in all vertebrate genomes for which data are available (Quina et al., 2012b). The 76 bp CE allele consists of a duplication within this core element. The genomic coordinates of the sequences shown are as follows: human, GRCh37/hg19 chr4:8,702,169-8,702,467; mouse, GRCm38/mm10, chr5:35,466,174-35,466,471; rat, Baylor 3.4/rn4, chr14:80,916,333-80,916,631; xenopus tropicalis, JGI 4.2/xenTro3, GL173219:238,403-238,694; cow, Baylor Btau_4.6.1/bosTau7, chr6:120,904,093-120,904,404. (C) Conserved transcription factor binding sites (top) and VISTA plot (bottom) of the Hmx1 CNE. Five TF binding sites for TF families of known developmental significance occur in this region; only those sites conserved across seven mammalian species are shown. In a DNA sequence of this length, the random expected occurrence of Ebox sites is two, of homeobox sites is two, and of the other sites is zero. VISTA plot shows percent homology between human and mouse genomes. The conserved transcription factor search was performed using Mulan: http://mulan.dcode.org/.