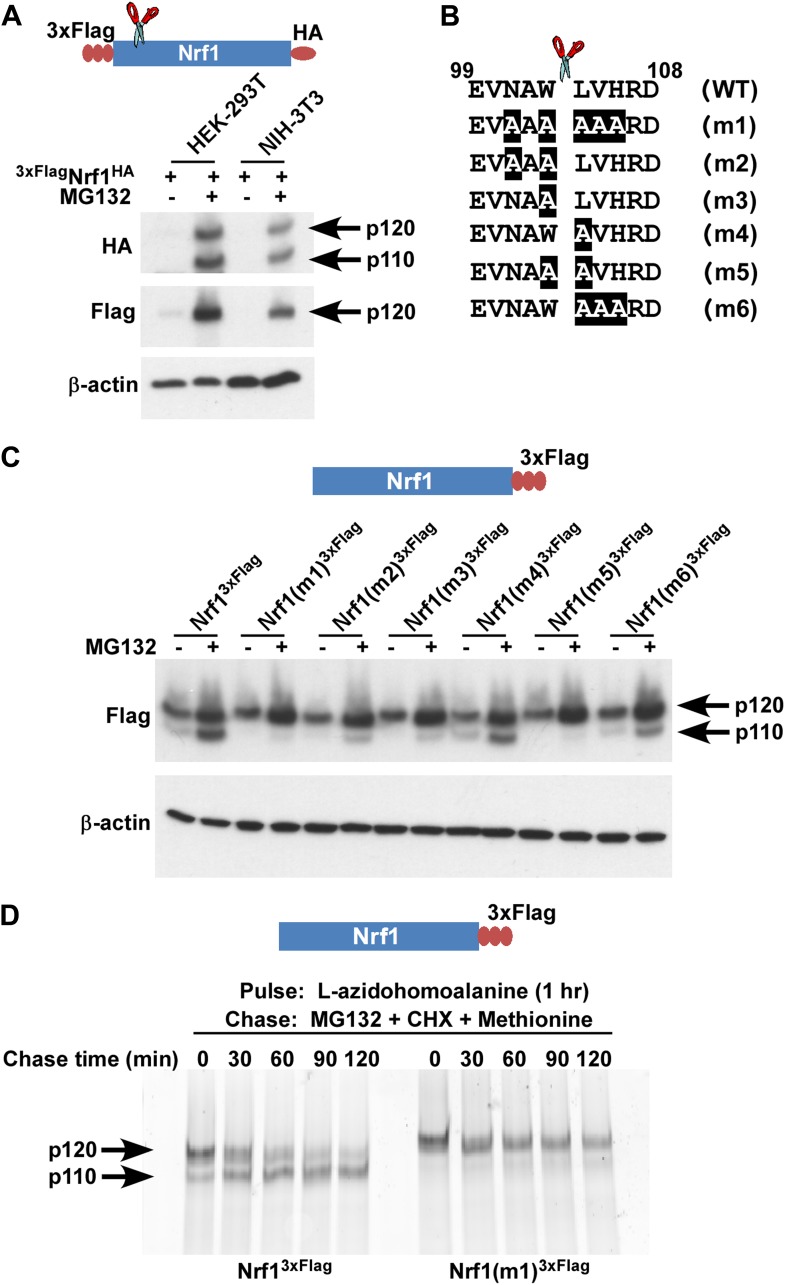
Figure 1. Nrf1 p110 is derived from p120 by proteolytic processing.
(A) Human HEK-293T or mouse NIH-3T3 cells were transduced with a retrovirus expressing 3xFlagNrf1HA and 72 hr later were left untreated or treated with 5 µM MG132 for 5 hr. The cell lysates were then used for immunoblotting with anti-Flag and anti-HA antibodies. β-actin protein levels were evaluated as a loading control. (B) Amino acids 99 to 108 from human Nrf1 are shown. The scissor indicates the cleavage position predicted by the Edman degradation sequencing. Also shown are mutants 1 through 6 with the mutated amino acids highlighted. (C) HEK-293T cells were transiently transfected with wild-type or various mutant constructs (m1 through m6) of Nrf13×Flag and 48 hr later were left untreated or treated with 5 µM MG132 for 5 hr. The cell lysates were then processed for immunoblotting with anti-Flag antibody. β-actin was used as a loading control. (D) HEK-293 cells stably expressing either wild-type Nrf13×Flag or Nrf1(m1)3×Flag were pulse-labeled for 1 hr with L-azidohomoalanine and then chased with MG132, cycloheximide (CHX), and excess methionine. The cells were harvested at the time points indicated (from 0 min to 120 min) and the lysates were subjected to immunoprecipitation with anti-Flag beads. Nrf1 species were visualized using the tetramethylrhodamine based click-chemistry method as described in ‘Materials and methods’.