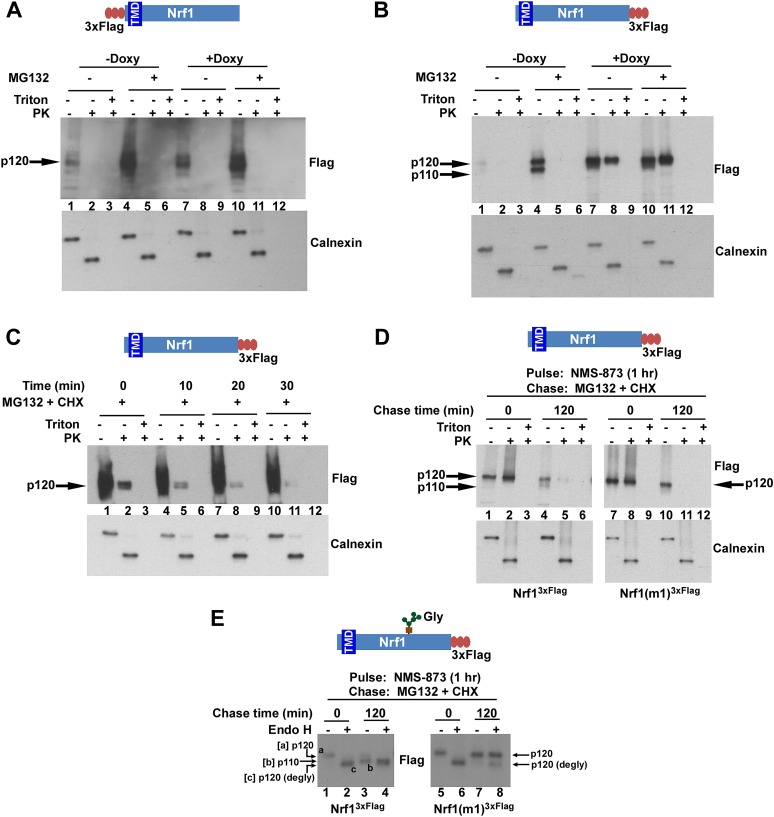
Figure 4. The C-terminus of Nrf1 is re-positioned from the lumen to the cytosol in a p97-dependent manner.
(A) HEK-293 cells stably expressing doxycycline (Doxy)-inducible shRNA targeting p97 (HEK-293-p97sh cells) were transiently transfected with a 3×FlagNrf1 expression construct and 24 hr later were incubated in the presence or absence of Doxy for 4 days after which the cells were further treated or not with 5 µM MG132 for 5 hr. Microsomes prepared from these cells were subjected to protease protection assay with Proteinase K (PK). SDS-PAGE followed by immunoblotting with anti-Flag antibody was used to detect Nrf1. Formation of a discrete cleavage product of Calnexin in the absence of detergent (Triton X-100) was used as an indicator for intact microsomes. (B) Same as (A), except that cells were transfected with an Nrf13×Flag expression construct. (C) HEK-293 cells stably expressing the non-cleavable Nrf1(m1)3×Flag were treated with MG132 plus cycloheximide (CHX) and cells were harvested at indicated time points. Microsomes from these cells were subjected to protease protection assay with PK. Samples were fractionated by SDS-PAGE followed by immunoblotting with anti-Flag antibody. (D) HEK-293 cells stably expressing either Nrf13×Flag or the non-cleavable Nrf1(m1)3×Flag were pulsed for an hour with 10 µM NMS-873 and then chased with MG132 plus cycloheximide (CHX). Microsomes prepared from cells harvested at the indicated time points (0 min and 120 min) were used in a protease protection assay with PK. SDS-PAGE followed by immunoblotting with anti-Flag antibody was used to detect Nrf1. (E) Total cell lysates from the pulse-chase experiment described above were subjected to deglycosylation by the enzyme Endoglycosidase H prior to SDS-PAGE and immunoblotting with anti-Flag. Species ‘a’ refers to full-length Nrf1 (p120) that was fully glycosylated. Species ‘b’ refers to Nrf1 that was proteolytically processed to p110 and degylcosylated following p97-dependent retrotranslocation. Species ‘c’ refers to species ‘a’ that was deglycosylated with Endo H. Note that species ‘b’ migrates slightly more slowly than species ‘c’ even though both species were deglycosylated and species ‘b’ lacked the N-terminal 103 residues. The unexpectedly slow mobility of species ‘b’ presumably reflects acquisition of additional post-translational modifications upon retrotranslocation on Nrf1’s CTD to the cytosolic side of the ER membrane. (TMD–transmembrane domain; Gly–N-linked glycan).
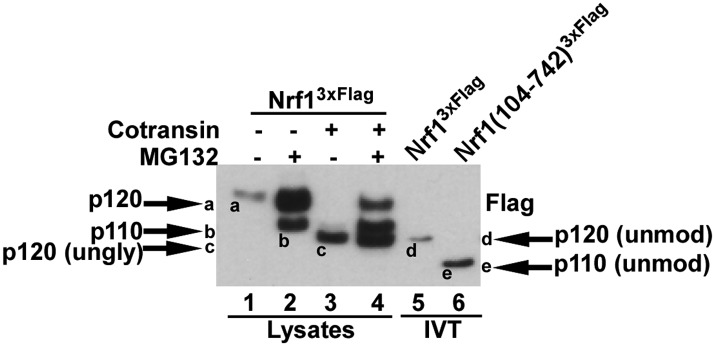
Figure 4—figure supplement 1. Different forms of Nrf1.