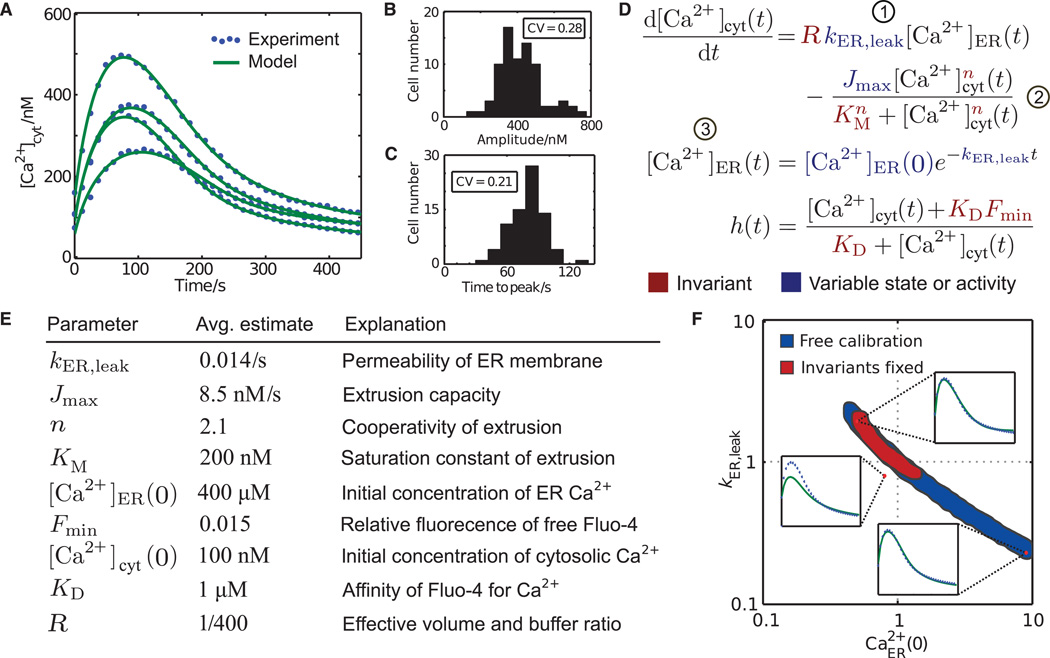
Fig. 2. Extraction of molecular parameter values from time-resolved, single-cell Ca2+ measurements.
(A) Single-cell Ca2+ responses upon addition of EGTA and thapsigargin. (B and C) Cell-to-cell variability of amplitude (B) and peak timing (C). CV, coefficient of variation. (D) Differential equations model that was used to infer parameter values. Numbers in circles denote terms illustrated in Fig. 1. (E) Population averages of single-cell parameter estimates. KD and R were linear with [Ca2+]cyt or [Ca2+]ER(0), respectively, and thus were always fixed (see Materials and Methods). (F) Exemplary slice through the seven-dimensional parameter space in which parameter sets were projected onto the plane spanned by the axes of ER leak (kER,leak) and the initial concentration of ER Ca2+ [Ca2+ER(0)]. Blue shading indicates the range of fold changes away from a simulated “true” parameter set in the center for which nearly identical fits could be obtained when all parameters were variable. Red shading shows the reduced range of parameters when the invariant parameters n, KM, and Fmin were constrained. The upper right and lower left insets show examples of parameter combinations that fit the same data. The upper left inset shows an example where this is not the case despite much smaller deviation from the true parameter set than the other two example parameter combinations.