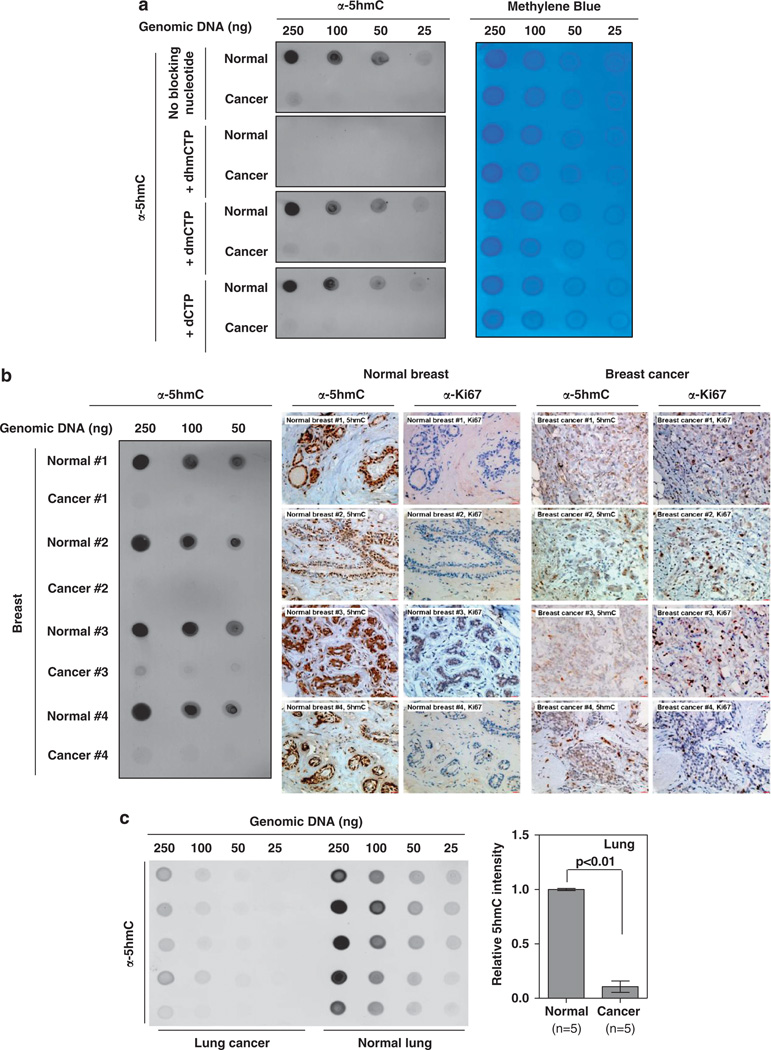
Figure 3.
Quantification of 5hmC decrease in human breast cancer. (a) To validate the dot-blot assay, the anti-5hmC antibody was preincubated with equal amount (1mm) of dhmCTP, dmCTP or dCTP for 1 h at room temperature, and then was used for dot-blot hybridization. Note that the anti-5hmC antibody very specifically recognized 5hmC, and could be effectively blocked by the preincubation with dhmCTP, but not dmCTP or dCTP. In addition, the dot-blot membrane was hybridized with 0.02% methylene blue in 0.3M sodium acetate (pH 5.2) to stain DNA. (b) Genomic DNAs were extracted from 15 pairs of paraffin-embedded human breast carcinoma and the matched normal breast tissues. In 12 of the 15 paired samples, sufficient genomic DNA was harvested for dot-blot hybridization. Representative micrographs of dot-blot assay, 5hmC and Ki67 staining in four paired samples are shown. Dot-blot results of the other eight pairs of normal and cancer samples are shown in Supplemental Figure S3. (c) Genomic DNAs were extracted from five pairs of paraffin-embedded human lung carcinoma and the matched normal lung tissues and subjected to dot-blot hybridization to determine the levels of 5hmC. Genomic DNAs were extracted from paraffin-embedded sections from normal and tumor samples following standard protocols. In detail, paraffin-embedded sections (10 ìm thickness) were deparaffinized by xylene. The deparaffinized samples were treated with ribonuclease A (50 µg/ml; Takara) at 37 °C for 1 h, and then incubated with proteinase K (0.3–0.5 mg/ml, Takara) in a digestion buffer (100mm NaCl/10mm Tris–HCl, pH 8.0 and 25mm EDTA, pH 8.0/0.5% SDS) at 55 °C overnight. After complete RNase and proteases digestion, genomic DNA was extracted with the same volume of phenol/chloroform/isoamyl alcohol (25:24:1) (Sangon, Shanghai), and then precipitated with an equal volume of isopropanol. After centrifugation, DNA pellet was washed once with 75% ethanol, air dried and dissolved in 15 µl distilled water. The DNA concentration was measured by NanoDrop (Thermo Scientific). The procedure for the dot-blot assay was modified from a procedure described previously (Xu et al., 2011). Briefly, DNA was spotted on a nitrocellulose membrane (Whatman), which was placed under an ultraviolet lamp for 20 min to crosslink the DNA. Subsequently, the membrane was blocked with 5% milk in TBS-Tween for 1 h and incubated with the primary anti-5hmC antibody at 4 °C overnight. After incubation with a horse radish peroxidase-conjugated secondary antibody anti-rabbit IgG (GenScript) for 1 h at room temperature, the membrane was washed with TBS-Tween 20 for three times and then detected and scanned by a Typhoon scanner (GE Healthcare). The 5hmC intensity was quantified by Image-Quanta software (GE Healthcare).