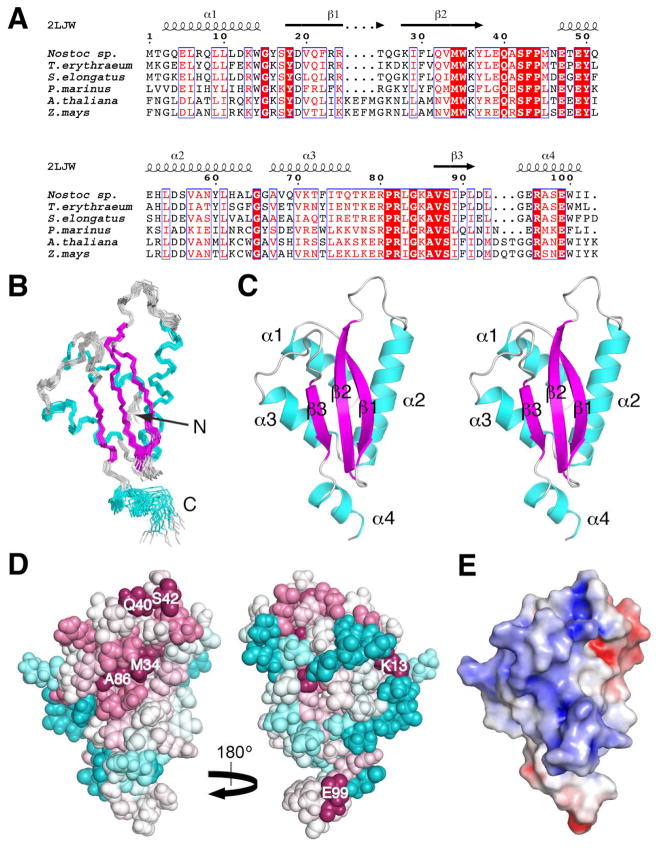
Fig. 1.
Solution NMR structure of Alr2454 from Nostoc sp. PCC 7120 (PDB ID, 2LJW). (A) Sequence alignment of a representative subset of the Pfam PF11267 protein domain family [1] from cyanaobacteria (Nostoc sp., Q8YUA0_NOSS1; Trichodesmium erythraeum, Q114L5_TRIEI; Synechococcus elongatus, Q31PB2_SYNE7; Prochlorococcus marinus, A2BVC5_PROM5, residues 4 to 105) and plants (Arabidopsis thaliana, Q8LDI1_ARATH, residues 113 to 222; Zea mays, B4FUD0_MAIZE, residues 116 to 225). Sequences were aligned using Clustal W 2.0 [33] and the sequence alignment was rendered using ESPript [34]. Boxed residues represent identical (white font, highlighted in red) and similar (red font) amino acid conservation. Residue numbering for Nostoc sp. Alr2454 and secondary structural elements [26] from its solution NMR structure (PDB ID, 2LJW) are drawn above the alignment. (B) Superposition of the final ensemble of 20 conformers of Alr2454 (residues 1 to 102). The view of the β-face of the protein is shown; α-helices and β-strands are shown in cyan and magenta, respectively, and loops are colored grey. (C) Stereoview of the lowest energy (CNS) conformer of Alr2454. Same orientation and color scheme as in (B). Secondary structural elements are labeled. (D) ConSurf [27, 28] image showing the conserved residues in Alr2454 (residues 3 to 101). Residue coloring, reflecting the degree of residue conservation over the entire PF11267 protein domain family (Pfam 25.0 [1]; 80 sequences), ranges from magenta (highly conserved) to cyan (variable). (Left) β-sheet face of the protein. (Right) α-helical face of the protein. Selected highly conserved residues are labeled. (E) APBS [29, 30] solvent accessible electrostatic surface potential of the β-sheet face of Alr2454 (residues 1 to 102) showing negative (red), neutral (white), and positive (blue) charges. All structure figures were rendered using PyMOL (www.pymol.org).