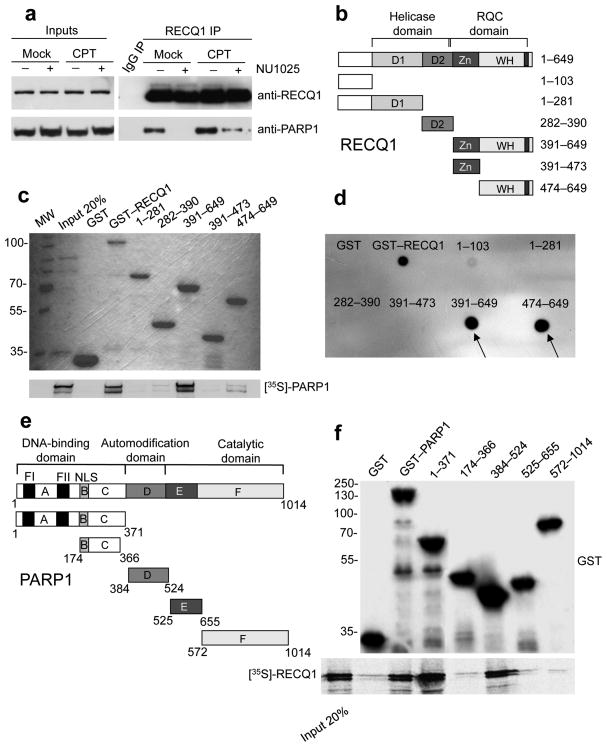
Figure 1. Analysis of the RECQ1-PARP1 interaction.
(a) IPs from U-2 OS cells using the anti-RECQ1 antibody ± PARP inhibitor (50 μM NU1025) and ± DNA damage (100 nM CPT for 2 hrs). (b) Schematic representation of the domain structure of RECQ1 and the GST-tagged RECQ1 fragments (D1 and D2 are the RecA-like domains). (c) Pull-down assays with GST-tagged RECQ1 fragments. Top: Coomassie stained gel of GST-RECQ1 fragments. Bottom: autoradiography of in vitro GST pull-down assay using 35S-labeled PARP1 protein. (d) Analysis of PAR binding in vitro. RECQ1 fragments (2 pmol) were dot-blotted onto a nitrocellulose membrane and incubated with 32P-labeled PAR. (e) Schematic representation of the domain structure of PARP1 and the GST-tagged PARP1 fragments (A, DNA binding domain; B, nuclear localization signal; D, BRCT–automodification domain; E, contains a WGR motif; F, catalytic domain. A third zinc-finger motif has been recently identified in domain C 36,37 in addition to the previously identified FI and FII zinc-finger motifs. NLS is a nuclear localization sequence. (f) Pull-down assays with GST-tagged PARP1 fragments. Bound proteins were revealed by autoradiography (bottom panel). Purified GST or GST–PARP1 proteins were detected with an anti-GST antibody (top panel). Input: 20% of the amount used in binding reactions.