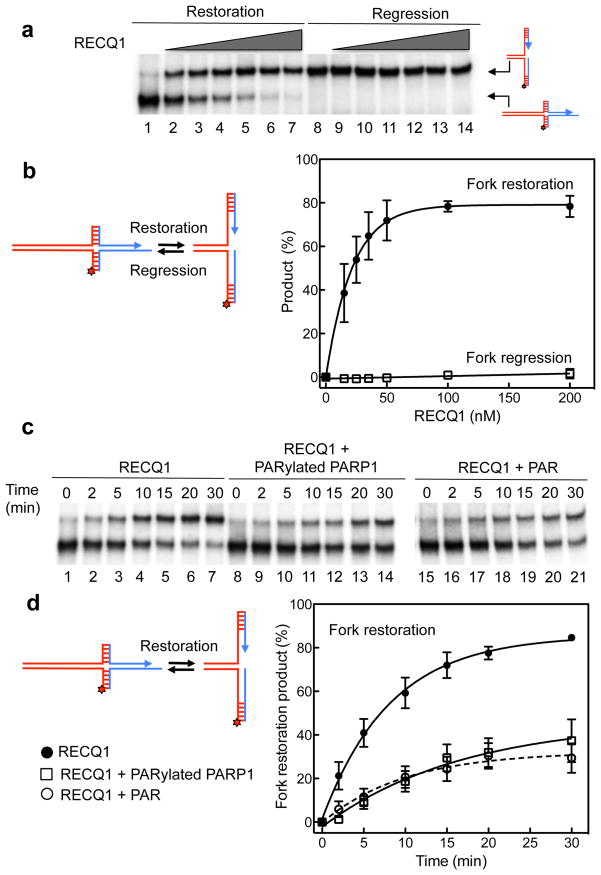
Figure 2. The in vitro fork restoration activity of RECQ1 is inhibited by PARylatedPARP1 and PAR.
(a) Lanes 1–7: fork restoration assays performed using increasing RECQ1 concentrations (0, 15, 25, 35, 50, 100, and 200 nM) and a fixed concentration of the chicken foot substrate (2 nM). Lanes 8–14: fork regression assays using increasing RECQ1 concentrations (0, 15, 25, 35, 50, 100, and 200 nM) and a fixed concentration of the replication fork structure (2 nM). The hatched regions indicate heterologous sequences that are included in the vertical arms to prevent complete strand separation. In addition, we inserted two mismatches and a single isocytosine modification to prevent spontaneous fork regression and restoration (see Supplementary Fig. 3 for more details). All the reactions were stopped after 20 min. (b) Left: reaction scheme. Right: Plot of the fork restoration and regression activities as a function of protein concentration. (c) Lanes 1–7: kinetic experiments performed using 40 nM RECQ1 and the chicken foot substrate (2 nM). Lanes 8–14: kinetic experiments performed in the presence of PARylatedPARP1 (40 nM). Lanes 15–21: kinetic experiments performed in the presence of PAR (100 nM). (d) Left: reaction scheme. Right: Plots of the fork restoration assays performed in the presence and absence of PARylatedPARP1 or PAR. The data points in a, b, c, and d represent the mean of three independent experiments. Error bars indicate standard error of the mean (s.e.m).