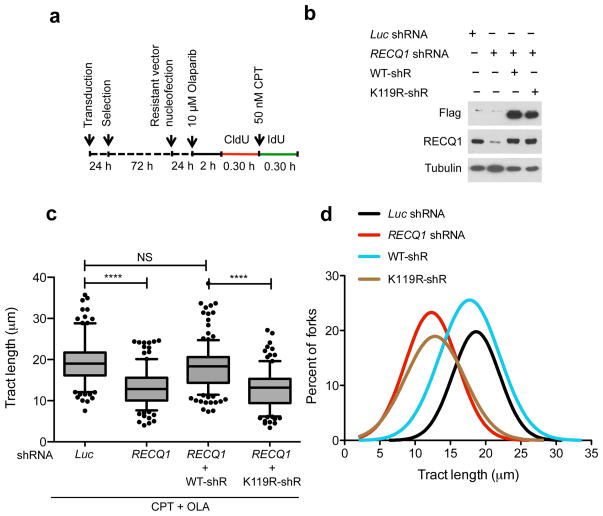
Figure 4. Genetic complementation of RECQ1-depleted cells with wild-type RECQ1, but not with the ATP-deficient K119R mutant, rescues the fork progression phenotype observed in RECQ1-depleted U-2 OS cells.
(a) Experimental scheme for the genetic knockdown-rescue experiments. U-2 OS cells were transduced with lentivirus to luciferase (Luc shRNA) or RECQ1 (RECQ1 shRNA). RECQ1-depleted cells were genetically complemented by nucleofection of RNAi resistant vectors, for the expression of wild-type (WT-shR) and ATPase-deficient (K119R-shR) shRNA resistant forms of RECQ1 before CldU labeling, as indicated. (b) Western blot analysis of RECQ1-depleted cells complemented with the shRNA resistant wild type RECQ1 or K119R mutant (both proteins are Flag-tagged). Tubulin was detected as a loading control. (c) Statistical analysis of IdU tract length measurements from a. Relative length of IdU tracts (green) synthesized after CPT treatment (50 nM). At least 175 tracts were scored for each dataset. 10 μM Olaparip was added 2 hours before CldU labeling and maintained during labeling. Whiskers indicate the 10th and 90th percentiles. Statistical test according to Mann-Whitney, results are ns not significant, **** p<0.0001. (d) The smoothened histogram shows green (IdU) tract length after Top1 and PARP inhibition in RECQ1-downregulated cells and RECQ1-downregulated cells genetically complemented with wild-type RECQ1 or the ATPase deficient K119R mutant.