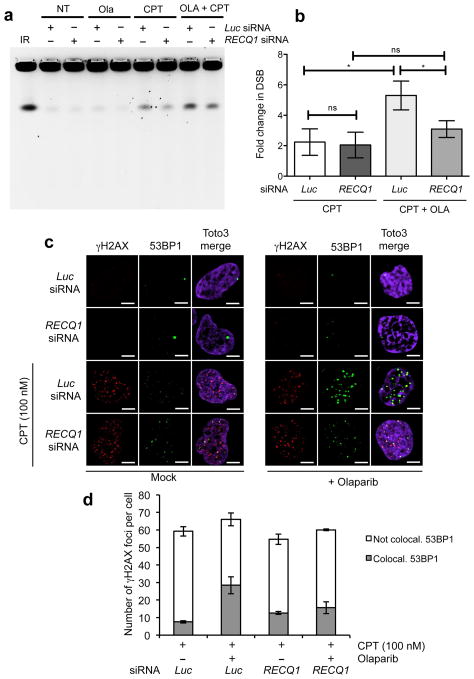
Figure 5. PARP inactivation leads to DSB formation at low CPT doses in the presence, but not in the absence of RECQ1.
(a) PFGE of U-2 OS cells transfected with siRNA against Luc and RECQ1, and treated with either CPT (100 nM) and/or Olaparib (10 μM). (b) DSB signals were quantified by ImageJ and normalized to unsaturated signals of DNA retained in the wells. The values obtained from the different treatments were then normalized against their respective untreated controls to obtain the fold change in DSBs upon treatment with CPT. The graph integrates results from three independent experiments. Statistical analysis was done using paired t test (* represents significance level where p value <0.05, ns represents non-significance between the samples) (c) Immunofluorescence experiments of U-2 OS cells were treated with 100 nM CPT, as indicated, ± 10 μM Olaparib, and co-stained for γH2AX and 53BP1. A representative image is shown in each condition. (d) The plot shows the average number of γH2AX foci per cell and the average fraction (± s.e.m.) of γH2AX foci colocalizing with 53BP1.