Figure 1. Schematic genome organization of Cowpea mild mottle virus (CpMMV) isolates identified in Ghana (top) and Florida (middle) as well as the general genome organization observed in carlavirus species (bottom; exact species shown in phylogenetic tree in Figure 2).
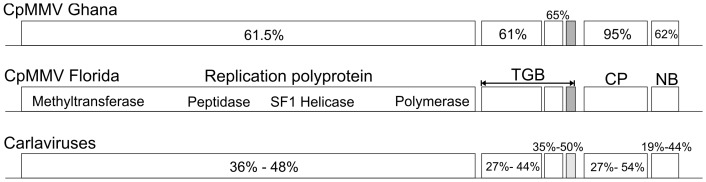
Each block represents identified open reading frames (ORFs) in these genomes and the encoded protein names are given on top of the CpMMV Florida panel, including the replication polyprotein, triple gene block (TGB), capsid (CP), and nucleic acid binding (NB) protein. The four different domains within the replication polyprotein exhibiting viral methyltransferase, peptidase, superfamily one (SF1) helicase, and RNA-dependent RNA polymerase (polymerase) motifs are indicated. Percentage values in the CpMMV Ghana genome schematic represent amino acid pairwise identities among individual ORFs of CpMMV Ghana and CpMMV Florida. Percentages in the Carlaviruses genome schematic represent amino acid pairwise identity ranges observed between different carlavirus species and CpMMV Florida. ORFs highlighted in grey represent ORFs without a standard start codon (a lighter grey in the carlaviruses panel indicates that some carlavirus species have a standard start codon while others do not).
