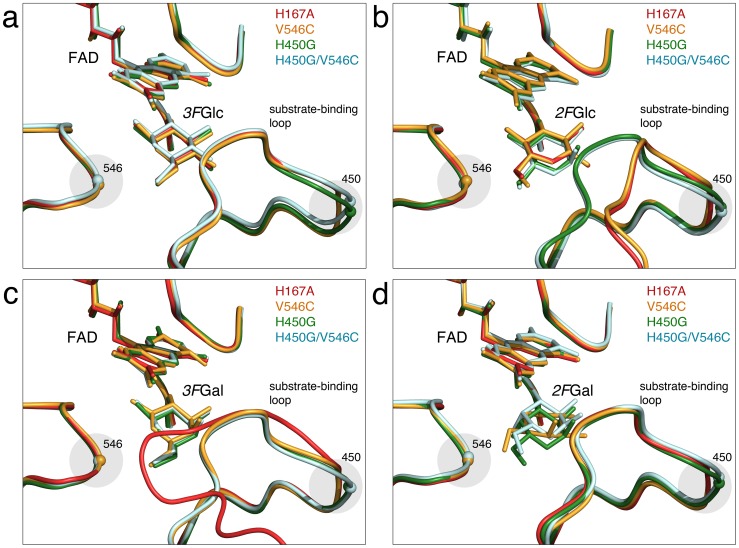
Figure 2. Comparison of conformation of the substrate-binding loop in TmP2O complexes with fluorinated sugars.
Superposition of mutant structures emphasizing the conformation of the substrate-binding loop. The FAD molecule and the pyranose sugar are shown as ball-and-stick models. (a) TmP2O variants complexed with 3FGlc corresponding to the productive 2-oxidation binding mode with the substrate-binding loop in the semi-open conformation. The relaxation induced by the H450G replacement (green and light-blue models) is highlighted by a shaded circle. The H167A model corresponds to PDB code 3PL8 [19]. (b) Mutant complexes with bound 2FGlc in the competing 3-oxidation binding mode. The H167A (PDB code 2IGO [14]) and V546C variants show the open loop conformation, which also has been observed for the wild type. H450G and H450G/V546C show the productive semi-open loop conformation. (c) Mutant complexes of V546C, H450G and H450G/V546C with bound 3FGal in the productive 2-oxidation binding mode with the substrate-binding loop in the semi-open conformation. The wild-type mimic H167A displays did not bind the sugar and displays the closed, occluded loop conformation that is typically observed for TmP2O in the absence of oxidizable sugar. The closed loop conformation is incompatible with sugar binding. (d) Mutant complexes with bound 2FGal. Despite the fundamentally different competing modes observed for H167A and V546C (C1-oxdiation mode) and H450G and H450G/V546C (C3-oxidation mode), all complexes show the substrate-binding loop in the semi-open loop conformation associated with productive sugar binding. The pictures were produced using the program PyMOL [43].