Fig. 1.
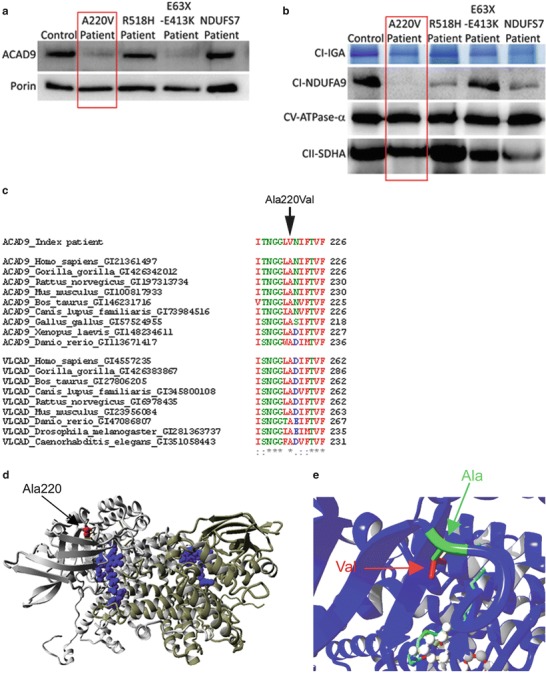
The ACAD9 p.Ala220Val substitution leads to instability of the protein. (a, b) ACAD9 expression level and complex I expression level in control, ACAD9 and NDUFS7 patient fibroblasts. The p.Ala220Val mutation is described in this chapter, whereas the two other ACAD9 patients (R518H and E63X-E413K) and the NDUFS7 patient were described previously (Nouws et al. 2010; Triepels et al. 1999). (a) SDS-PAGE immunodetections of ACAD9 and porin, showing a decreased amount of ACAD9 in the patient. (b) BN-PAGE analysis of complex I in-gel activity (CI-IGA) and western blot immunodetection of the complex I subunit NDUFA9, showing a decreased activity and amount of fully assembled complex I in the patient. Complex V-CV-ATPase-α and complex II-SDHA were used as loading controls. (c) Conservation of the altered amino acids is shown via clustalW alignments. Asterisks (*) indicate identical amino acids, colons (:) indicate conserved substitutions, and periods (.) indicate semiconserved substitutions. (d) Model of ACAD9 (PDB code 3b96) and the p.Ala220Val mutation, showing the two ACAD9 monomers (grey and yellow), the location of the FAD cofactor (blue), and the arrow indicates the location of the mutation (red). (e) The mutation shown in closer detail; the replacement of alanine (in green and indicated by green arrow) by valine leads to an addition of two methylgroups (indicated in red and by a red arrow). Moreover, the mutation is in proximity to the fatty acid (light blue stretch) and FAD (white balls) binding sites in the core of the protein
