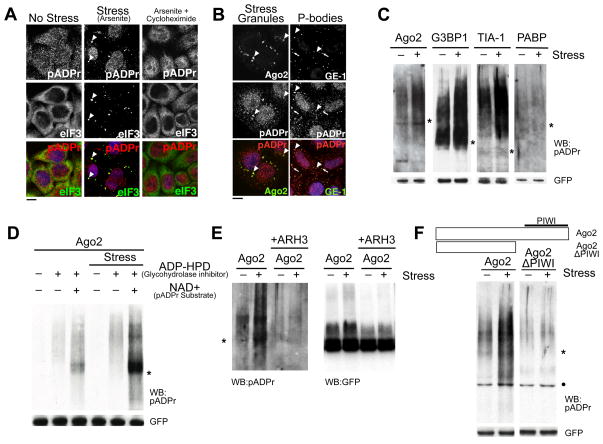
Figure 1. pADPr is enriched in SGs upon multiple types of stresses and modifies specific cytoplasmic RNA-binding proteins dependent on RNA-binding domain.
(A) pADPr staining using LP96-10 antibodies in HeLa cells untreated, or treated with 100 μM arsenite for 60 min, or for 30 min followed by 100 μM arsenite +100 μg/ml cycloheximide for 30 min. Arrowheads, SGs; scale bar, 10 μm. (B) HeLa cells treated with 100 μM arsenite were stained for pADPr, SG component Ago2 (arrowheads) or PB component GE-1 (arrows). DNA was stained with Hoeschst 33342 (blue); scale bar, 10 μm. (C) Immunoprecipitates of 4 GFP-tagged SG-localized RNA-binding proteins from cells treated with or without 20 nM pateamine A were probed for pADPr. (D) Immunoprecipitates of GFP-Ago2 from cells treated with or without 20 nM pateamine A were probed for pADPr. The cell extracts either included or excluded 1 μM ADP-HPD, and with or without 1 mM NAD+ before immunoprecipitation by anti-GFP. (E) pADPr modification of Ago2 from cells treated with or without 20 nM pateamine A was verified by treating the immunoprecipitates with ARH3. The immunoprecipiates were probed for pADPr (left) and GFP (right). (F) Immunoprecipitates of wild-type and PIWI mutant of GFP-Ago2 from cells treated with or without 20 nM pateamine A were probed for pADPr. For panels C-F, cell extracts included 1μM ADP-HPD unless stated otherwise; shown are western blots for pADPr (LP96-10) and GFP levels in each immunoprecipitate. Asterisks indicate the position of the corresponding GFP-tagged RNA-binding protein constructs. Black dots indicate non-specific binding to BSA by LP96-10. See also Figure S1.