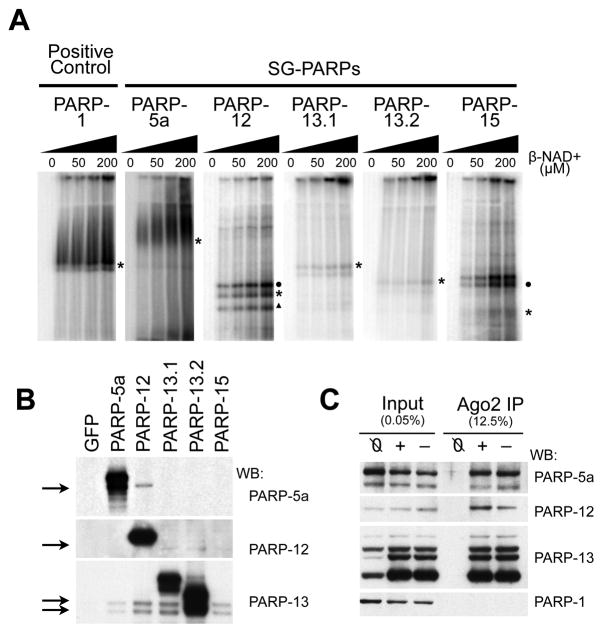
Figure 4. PARP-13 family members are poly(ADP-ribosylated) by other SG-PARPs.
(A) in vitro ADP-ribosylating assay for PARP-1, -5a, -12, -13.1, -13.2 and -15. HeLa S3 cells were transfected with individual GFP-tagged SG-PARPs and GFP-PARP immunoprecipitates were washed twice with 450 mM NaCl then once with 150 mM NaCl. The immunoprecipitates were incubated with 0, 25, 50, 100 or 200 μM NAD+ (with 1/175 fold of P32-labeled NAD+) at 16°C for 30 min, separated on a 6% SDS-PAGE gel and visualized by autoradiography. Asterisks indicate the position of the corresponding GFP-tagged SG-PARPs; circles and triangle indicate the endogenous position of PARP-13.1 and PARP-13.2 respectively. (B) Western blots of the immunoprecipitates from panel A were probed with PARP-5a, -12 and -13 (antibodies for PARP-15 are not good for detecting endogenous protein). Immunoprecipitates from cells transfected with GFP were used as a negative control. (C) HeLa S3 cells were transfected with GFP-tagged Ago2 and cells either treated with (+) or without (−) 20 nM pateamine A for 30 min. Untransfected cells treated with 20 nM pateamine A were used as a negative control (Ø). The cytoplasmic lysates were immunoprecipitated with anti-GFP and washed thrice with cytoplasmic lysis buffer. The input and immunoprecipitates were probed with antibodies against PARP-1, -5a, -12 and -13. See also Figure S4.