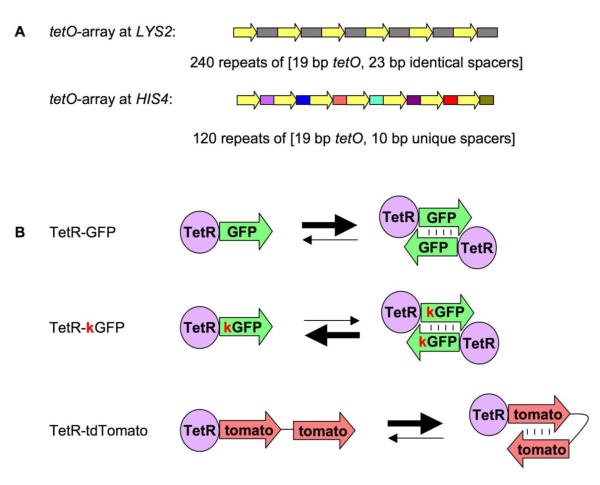
Figure 1. Arrays and proteins used in this study.
A. Arrays used in this study.
The two arrays used in this study are: tetO array at LYS2 locus and tetO array at HIS4 locus. The former consists of 240 copies of a DNA fragment, which contains 19 bp tetO binding site and a 23 bp spacer (thus all spacers in the array are identical). The latter consists of 120 DNA fragments, each one of which contains the 19 bp tetO binding site and a unique 10 bp spacer (thus all spacers in the array are different).
B. Proteins used in this study.
The three proteins used in this study are: TetR-GFP, TetR-kGFP and TetR-tdTomato. TetR-GFP has a strong tendency to form inter-molecular dimers due to GFP dimerization. TetR-kGFP carries A206K mutation, which disrupts this property. TetR-tdTomato has two units of Tomato attached to TetR, thus it can form intra-molecular dimers instead of inter-molecular dimers.