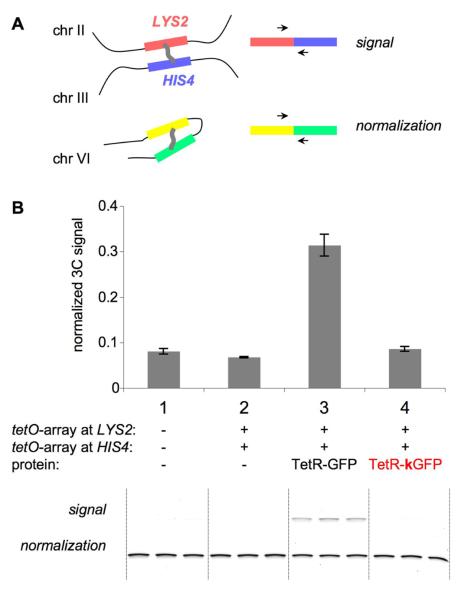
Figure 2. 3C assay of spatial proximity between chromosomal loci.
A. Schematic representation of the 3C assay.
3C assay relies on chromatin crosslinking and subsequent PCR analysis of ligation junctions between crosslinked segments. Signal PCR assays spatial proximity of LYS2 locus on chromosome II and HIS4 locus on chromosome III, where tetO arrays are integrated. Normalization PCR, which assays spatial proximity between two randomly chosen chromosomal segments located in cis, is used to control for the efficiency of the procedure in all samples.
B. 3C assay of spatial proximity between HIS4 and LYS2 loci.
Below – gel (reactions are done in triplicates, such that 3 PCR reactions were set up using 1 3C template), above – quantification of gel products. Case 1 – control WT strain without arrays and repressor proteins. Cases 2, 3, 4 – tetO arrays are present at HIS4 and LYS2 loci. Case 2 – no repressor protein, case 3 – TetR-GFP, case 4 – TetR-kGFP.
p(1,2) = 8.64 x10−2; p(1,3) = 3.44 x10−3; p(1,4) = 4.09 x10−1; p(2,3) = 4.60 x10−3; p(2,4) = 3.4 x10−2; p(3,4) = 3.92 x10−3 (all p-values that include sample 3 are < 0.01; underlined).