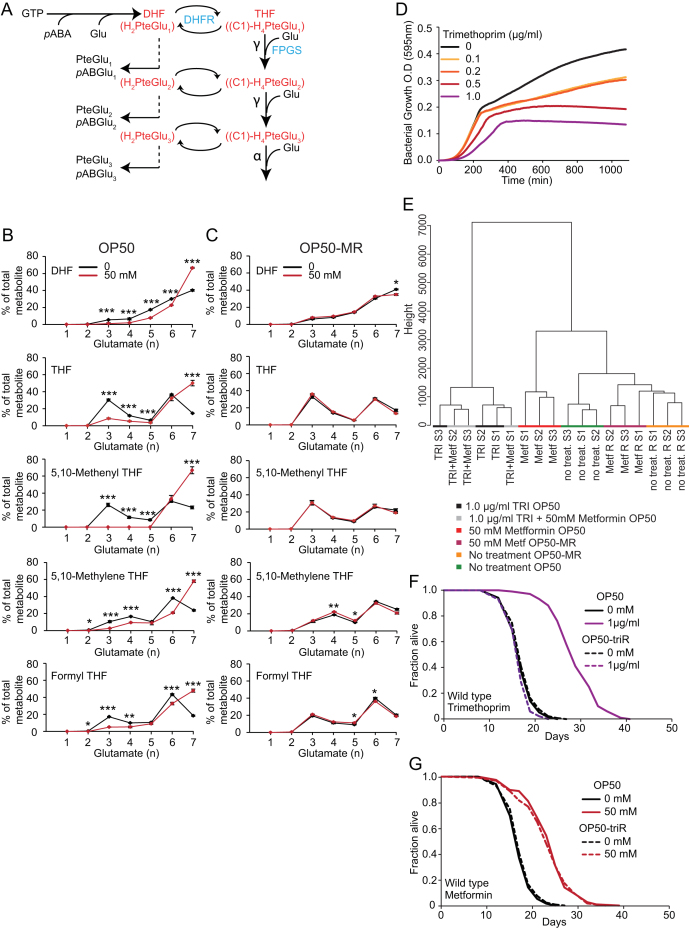
Figure S4.
Metformin and Trimethoprim Alter Bacterial Metabolism and Extend C. elegans Lifespan by Common Mechanisms, Related to Figure 4
(A) Diagram showing folate synthesis, one-carbon substitution, polyglutamylation and catabolism in E. coli (adapted from Kwon et al., 2008). Blue, enzymes involved in these reactions: FPGS, folylpolyglutamate synthetase; DHFR, dihydrofolate reductase. Red: DHF, dihydrofolate; THF, tetrahydrofolate. Black: pABA, p-aminobenzoic acid; Glu, glutamate; GTP, guanosine triphosphate; PteGlu, pteroylmonoglutamic acid; pABGlu, p-aminobenzoyl-l-glutamate.
(B) Folate polyglutamylation profiles of detectable folate metabolites of E. coli OP50 grown in the presence or absence of metformin. Metformin strongly affects polyglutamylation levels of all folates detected.
(C) Folate polyglutamylation profiles of detectable folate metabolites of E. coli OP50-MR bacteria grown in the presence or absence of metformin (note the absence of effects in most cases).
(D) Trimethoprim (TRI) acts as a bacteriostatic antibiotic to delay bacterial growth in a dose-dependent manner.
(E) Hierarchical cluster analysis of metabolites of TRI and metformin-treated OP50 and OP50-MR using the Ward method. Metabolite clusters from the TRI+metformin condition or TRI alone are indistinguishable from each other. Metformin treatment of OP50-MR creates a cluster of metabolites similar to OP50-MR and OP50 in the absence of treatment and distinct from OP50 treated with metformin.
(F) TRI does not extend worm lifespan in the presence of a TRI-resistant strain of E. coli OP50 (OP50-triR) expressing the type IIa DHF reductase (Kim, 2009).
(G) TRI-resistance in E. coli has no effect on induction of life extension by metformin.
For statistics see Table S4. Error bars, SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.