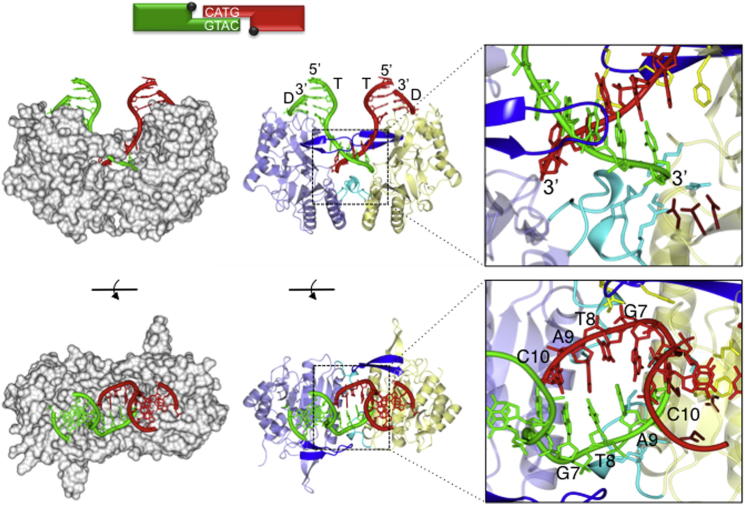
Figure 2.
Architecture of an Annealed dsDNA Break Bound to an NHEJ Polymerase
Schematic representation of the annealed DNA double-strand break present in the crystal structure with the annealed microhomology sequence highlighted. Below this scheme are representations of the crystal structure of the annealed DNA double-strand break bound to an NHEJ polymerase, PolDom. The figure depicts a synaptic complex formed between two binary (DNA [T/D]-PolDom) complexes that have come together, in a “face-to-face” orientation, by annealing of the 3′ self-complementary DNA overhangs of the break. To the left of the figure, the polymerase is depicted as a gray solvent accessible surface, and the DNA is depicted in red or green (side-on and top-down views). The polymerases facilitate DNA break synapsis between discontinuous DNA ends by cradling the termini, within a continuous molecular surface, promoting microhomology-mediated end synapsis.
The middle of the figure has a protein ribbon representation of the structure of the annealed DNA break bound to PolDom (side-on and top-down views). The polymerase monomers are colored light blue and yellow, respectively. Significant structural elements loop 1 and loop 2 are colored blue and cyan, respectively. The polymerase induces a major splaying (∼119°) of the template strand (T). The resulting 3′ overhangs are annealed together, forming four Watson-Crick base pairs (G7-C10), via a region of microhomology. This end synapsis is promoted by interactions with loops 1 and 2 (inset). The template strand from one binary complex terminates in the active site of the opposing binary complex, effectively becoming an incoming primer strand (inset). The inset also features catalytic site residues (tan) as well as residues involved in template strand splaying and orientation (yellow) and primer strand orientation and tethering (cyan) (see Figures 3A, 6A, 6B, 7A, 7B, and S4–S7 for more detail).
See also Figure S2.