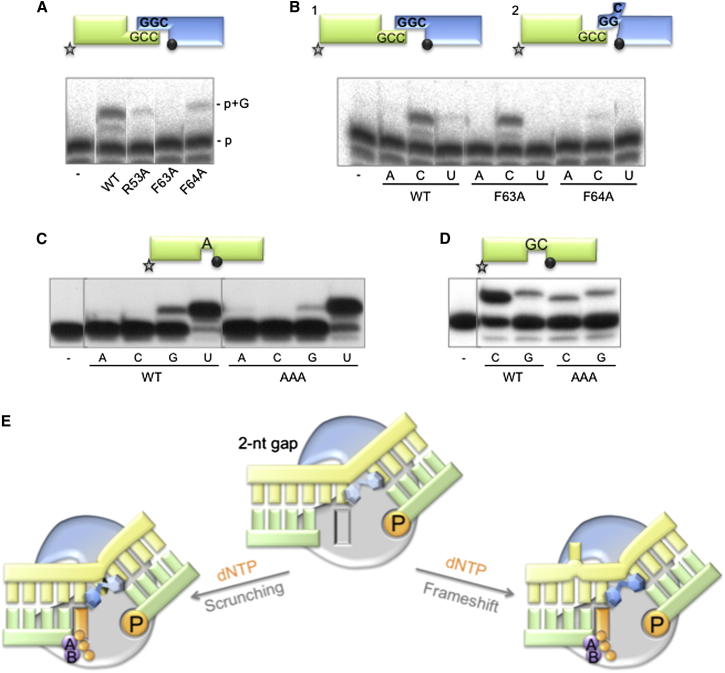
Figure 5.
Selecting the Templating Base: Roles of Residues Phe63 and Phe64
(A and B) NHEJ reactions were performed with 600 nM of the indicated proteins using a set of DNA substrates formed by hybridizing the oligonucleotides CCG with NHEJ-D and GGC with NHEJ-D2. In (A), only GTP (100 μM) was added in the presence of 1 mM MnCl2, whereas in (B) the other three nucleotides were added (100 μM).
(C and D) Gap-filling reactions were performed as described in Experimental Procedures for the indicated proteins (25 nM) using a gapped DNA substrate containing the oligonucleotides SP1C, T13C, and DG-P (C) or P15, T17, and DG2P (D). When indicated, NTPs were added separately at 10 nM in the presence of 1 mM MnCl2.
(E) A cartoon showing the dichotomy that PolDom confronts when dealing with gaps longer than 1 nt during NHEJ; the template strand is either “scrunched,” and the gap filled in correctly (left side), or the template strand is dislocated and sequence is lost with the production of frameshifts (right side). The protein is shown as a gray surface with a blue section indicating the approximate position of loop 1, 5′P and incoming nucleotide are colored orange, the two metal ions are shown in purple, and the DNA substrate is shown in yellow (template strand) and green (primer and downstream strands). Phenylalanines Phe63 and Phe64 are shown as blue hexagons holding the kink in the DNA substrate, indicating with a darker blue color their importance for each reaction. See main text for details.