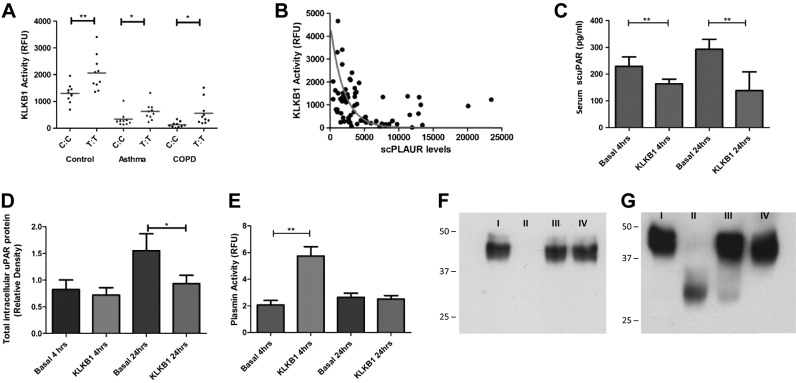
Figure 5.
SNP rs4253238 modulates KLKB1 enzymatic activity, which, in turn, determines scuPAR levels. A) KLKB1 activity assay identified an elevation of KLKB1 activity with the rs4253238 T:T genotype in control (P=0.004), asthma (P=0.034), and COPD (P=0.011) populations. KLKB1 activity was also found to be reduced in the 2 diseased populations when compared to controls (P=1×10−4). B) KLKB1 activity was found to be inversely correlated with scuPAR levels regardless of whether the subjects were controls, patients with asthma, or patients with COPD (P=1×10−4; R2=0.278). C, D) Exposure of NHBECs to plasma-extracted human KLKB1 results in reduced levels of scuPAR in cell supernatants at 4 and 24 h (P=0.015 and P=0.029, respectively; C), while total intracellular protein was decreased at 24 h (P=0.05; D). E) Within the same cell system, plasmin was activated by KLKB1 at 4 h, but not at 24 h (P=5×10−3), identifying the need for a cell-free system to investigate the direct effect of KLKB1 on scuPAR. F, G) Staining of recombinant human uPAR (ruPAR) digested with KLKB1 with the uPAR DI-specific monoclonal antibody IIIF10 (F) and the polyclonal antibody BAF 807 (G) reveals that KLKB1 proteolytically cleaves the ruPAR molecule at the DI region (F), leaving behind a fragment of ∼30 kDa, comparable to that formed following digestion of a known uPAR proteolytic enzyme, chymotrypsin (P<0.001). I, ruPAR; II, ruPAR + KLKB1; III, ruPAR + chymotrypsin; IV, ruPAR + KLKB1 + protease inhibitor cocktail. *P < 0.05, **P < 0.001.