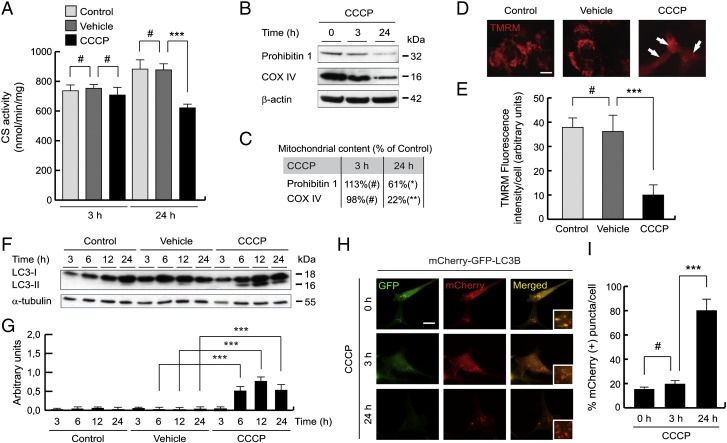
Fig. 1.
Mitochondrial damage and autophagy induction in SH-SY5Y CCCP-treated cells. (A) SH-SY5Y cells were exposed with 10 μM CCCP, with vehicle (0.05% (v/v) ethanol) or without any treatment (control), lysed and CS activity measured. Data are expressed as nmol/min/mg protein (#p > 0.05; ***p ≤ 0.001). (B and C) SH-SY5Y cells were treated with 10 μM CCCP for 0, 3 or 24 h and lysates separated by SDS-PAGE and Western-blotting performed. Blots were probed with antibodies against two inner mitochondrial membrane proteins, COX IV and prohibitin 1. β-actin was used as a loading control. (B) Representative blot of at least three independent experiments. (C) Densitometry of each band expressed as % of control (#p > 0.05; *p ≤ 0.05; **p ≤ 0.01). (D and E) SH-SY5Y cells were exposed with 10 μM CCCP, with vehicle (0.05% (v/v) ethanol) or without any treatment (control) for 24 h, and stained with TMRM to assess Δψm by immunofluorescence. (D) Representative microphotographs of TMRM stain. Scale bar represents 10 μm. The arrows highlight cells with low Δψm. (E) TMRM fluorescence intensity per cell (in AU) by immunofluorescence (# p > 0.05; *** p ≤ 0.001). (F and G) SH-SY5Y cells were exposed with 10 μM CCCP, with vehicle (0.05% (v/v) ethanol) or without any treatment (control), harvested by trypsinization at different times and lysed. The ratio LC3-II/LC3-I was determined by Western-blotting. α-tubulin expression was used as a loading control. (F) Representative blot of at least three independent experiments. (G) Densitometry of each band expressed in arbitrary units of intensity (***p ≤ 0.001). Molecular mass is indicated in kilodaltons (kDa) next to the blots. Data were expressed as mean ± SEM; n = 3. (H and I) SH-SY5Y cells were transfected with mCherry-GFP-LC3B plasmid for 24 h and treated with 10 μM CCCP for 0, 3 or 24 h and fixed. (H) Representative immunofluorescence microphotographs. Autophagolysosomes and autophagosomes were labeled by red (mCherry-LC3B) and yellow puncta (mCherry-GFP-LC3B), respectively. The boxes highlight the pattern of each condition. (I) Percentages of mCherry (+) puncta per cell (#p > 0.05; ***p ≤ 0.001). Scale bar represents 10 μm.