Figure 2.
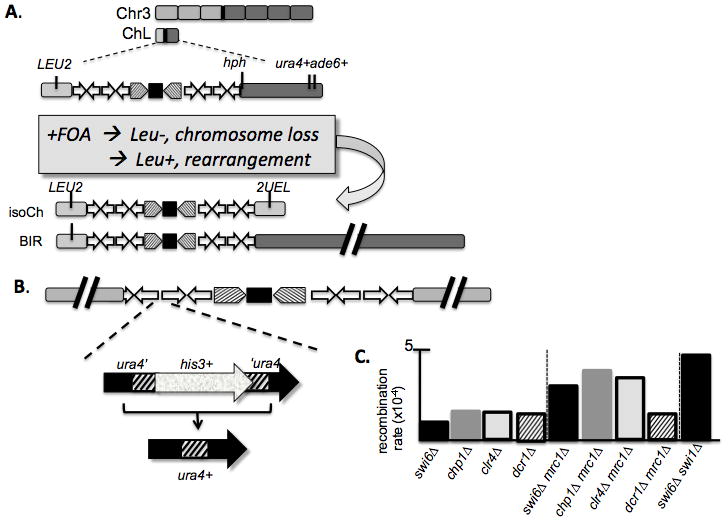
A. Minichromosome model for chromosome rearrangement [Tinline-Purvis, 2009 #5381][Nakamura, 2008 #5382] leads to isochromosome formation or acrocentric deriiatives through BIR. B. Internal model for centromere rearrangement uses an interrupted ura4+ gene to monitor recombination. C. Double mutants that disrupt Swi6 or Chp1 and components of replication fork stability have enhanced defects, using the system in Fig 2b. Median recombination rate reported. Original data and statistical analysis from [Li, 2013 #6465].
