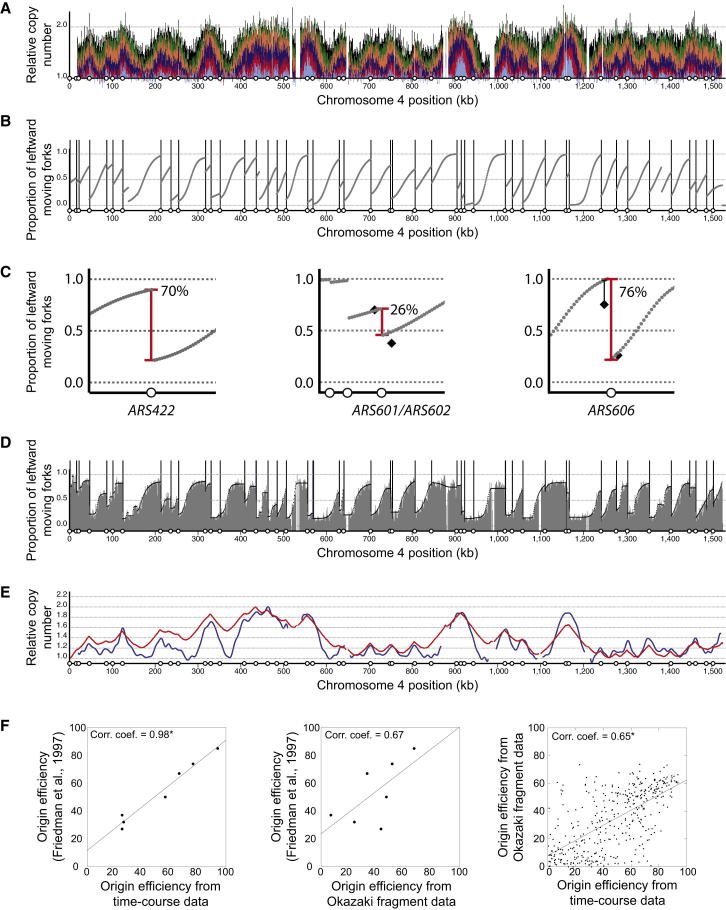
Figure 3.
Genome-wide Replication Fork Direction and Origin Efficiency from Time-Course Data
(A) Replication profile for chromosome 4 (as described for Figure 1A).
(B) Proportion of leftward-moving forks across chromosome 4 inferred from replication-time-course data. Vertical lines mark the location of active origins, at each of which there is a sharp transition from leftward- to rightward-moving forks.
(C) Proportion of leftward-moving forks for 50 kb regions centered on ARS422 (left), ARS601/ARS602 (center), and ARS606 (right). Red vertical lines indicate the magnitude of the transition from leftward- to rightward-moving forks and the efficiency of the origin. Black diamonds indicate the proportion of leftward-moving forks at four chromosome 6 locations as previously determined by fork-direction gels (Friedman et al., 1997); for the location to the left of ARS606, the value was determined to be >75% and origin activity was estimated to be 74%.
(D) Proportion of leftward-moving forks across chromosome 4 derived from mapping Okazaki fragments. Grey shading indicates the raw data with a fitted curve shown in black (origin locations are marked as in [B]; Smith and Whitehouse, 2012).
(E) Replication time expressed as relative copy number derived from Okazaki fragment data (red; Smith and Whitehouse, 2012) or directly by deep sequencing (blue; Müller and Nieduszynski, 2012).
(F) Pairwise comparisons of origin efficiency determined from the time-course data, from fork-direction gels (Friedman et al., 1997), and from mapping Okazaki fragments (Smith and Whitehouse, 2012). Pearson correlation coefficients are given (∗p < 0.0001).