Abstract
Traumatic brain injury (TBI) is a major cause of death and disability worldwide. Robust, reproducible segmentations of MR images with TBI are crucial for quantitative analysis of recovery and treatment efficacy. However, this is a significant challenge due to severe anatomy changes caused by edema (swelling), bleeding, tissue deformation, skull fracture, and other effects related to head injury. In this paper, we introduce a multi-modal image segmentation framework for longitudinal TBI images. The framework is initialized through manual input of primary lesion sites at each time point, which are then refined by a joint approach composed of Bayesian segmentation and construction of a personalized atlas. The personalized atlas construction estimates the average of the posteriors of the Bayesian segmentation at each time point and warps the average back to each time point to provide the updated priors for Bayesian segmentation. The difference between our approach and segmenting longitudinal images independently is that we use the information from all time points to improve the segmentations. Given a manual initialization, our framework automatically segments healthy structures (white matter, grey matter, cerebrospinal fluid) as well as different lesions such as hemorrhagic lesions and edema. Our framework can handle different sets of modalities at each time point, which provides flexibility in analyzing clinical scans. We show results on three subjects with acute baseline scans and chronic follow-up scans. The results demonstrate that joint analysis of all the points yields improved segmentation compared to independent analysis of the two time points.
Keywords: Image segmentation, Atlas formation, Longitudinal analysis
1. INTRODUCTION
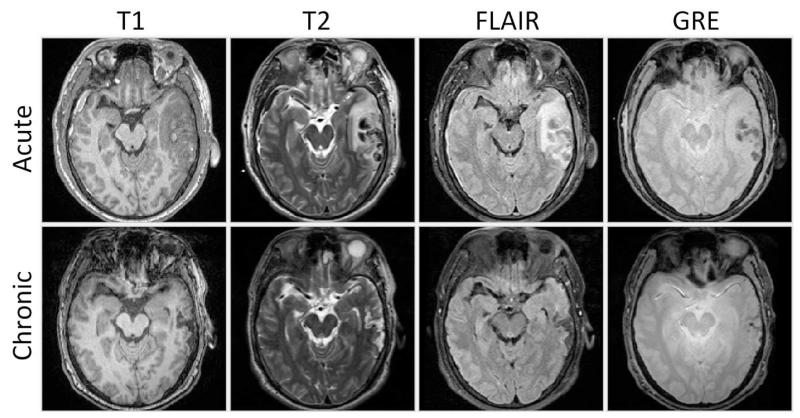
Traumatic brain injury (TBI) occurs when an external force traumatically injures the brain, typically due to car accidents, accidental falls, and wartime injuries. It is a major cause of death and disability worldwide, especially in children and young adults, and it affects 1.7 million Americans annually.1,2 Robust, reproducible segmentations of MR images with TBI are crucial for quantitative analysis of recovery and treatment efficacy. However, this is a significant challenge due to severe changes such as edema (swelling), bleeding, tissue deformation, skull fracture, and other effects related to head injury. Acute and chronic images of Subject I is shown in Fig. 1 where non-hemorrhagic lesions (edema / swelling) are shown as hyperintense regions in FLAIR while hemorrhagic lesions (bleeding) are shown as hypointense regions in T2 and GRE. Despite the clinical importance of quantifying changes in TBI patient images, few research has been done on segmentation of MR images of TBI patients. A previous work by Thatcher et al.3 used fuzzy C-means and/or k-nearest-neighbor (kNN) algorithms and manual classification to segment 3D MR images of TBI patients without a longitudinal component.
Figure 1.
Axial views of acute and chronic images of Subject I.
Many others have proposed automatic segmentation methods for 3D MR images. For segmenting normal brain MR images, van Leemput et al.4 and Zhang et al.5 proposed atlas based methods, while Tu et al. proposed a learning-based method.6 Brain tumor is an example of a pathology that have similar properties to TBI. For segmenting brain MR images with tumor, Prastawa et al. proposed methods based on outlier detection7 and subject specific modification of atlas priors.8 Clark et al. proposed a automatic tumor segmentation using knowledge-based techniques.9 Ho et al. proposed a level-set based tumor segmentation method.10 Menze et al. presented a generative model for brain tumor segmentation using multi-modal MR images.11 For brain MR images with TBI, automatic segmentation is difficult due to the variability of lesion types, shapes, and appearances. In this paper, we introduce a novel user-initialized multi-modal image segmentation framework for longitudinal (4D) MR images with TBI.
This paper is organized as follows: Section 2 describes a patient-specific segmentation framework for longitudinal MR images of TBI patients. The framework is composed of Bayesian segmentation with user initialization, and personalized atlas construction. Section 3 shows the results of the proposed method. We compare the result of temporally independent segmentation with our result. We present conclusions and potential future work related to our method in Section 4.
2. METHOD
2.1 Bayesian segmentation with user initialization
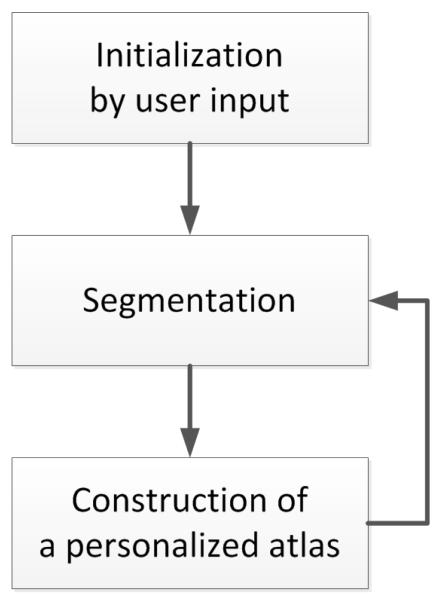
The segmentation framework for multi-modal MR images is depicted in Fig. 2. The framework is initialized through manual input of primary lesion sites and affine-registered atlas at each time point, which are then refined by a joint approach composed of Bayesian segmentation and construction of a personalized atlas.
Figure 2.
Our semi-automatic segmentation framework for MR images with TBI.
Suppose the multi-modal images at time point t are Xt = x1,t, … , xN,t with Dt the number of channels, where we use mixtures of Gaussians to model the data following van Leemput et al.4 We segment the images by maximizing the log likelihood function:
| (1) |
where t ∈ {1 … T } is the number of time points, Kt is the number of classes at time point t, is the multivariate Gaussian distribution, αi,j,t = p(zi,t = j) is the prior, and zi,t ∈ {1 … K} is the tissue class label at position i and time point t. We use the Expectation-Maximization (EM) algorithm12,13 to maximize the log likelihood function.
E step: update the expectation weights p(zi,t = j|xi,t, μj,t, Σj,t) at each location i of each time point t.
M step: update the Gaussian parameters μj,t, Σj,t at time point t.
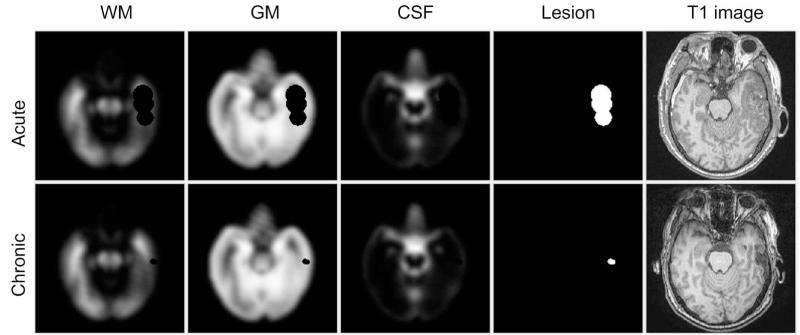
We use user input and an affine-registered atlas to initialize the parameters μj,t, Σj,t, and αj,t. The user input are spheres St indicating lesions and the number of lesion classes Lt at each time point t. We use the K-means algorithm14 to get initial estimates μj,t and Σj,t for each lesion class. For other tissue classes (white matter (WM): k = 1, gray matter (GM): k = 2, cerebrospinal fluid (CSF): k = 3 and background (BG): k = 4), we use the affine-registered atlas (masked by user input St) to estimate μj,t and Σj,t. The initial priors αj,t of each class are obtained by modifying the standard atlas using St, following Prastawa et al.7 We assume that lesions are found in WM and GM regions, so the initial prior for each lesion class becomes αj,t = w(α1,t + α2,t)+St, for j ≥ 5, where w is a uniform weight for lesions chosen to be 0.001 in our calculation. The αj,t of other classes are linearly transformed to ensure that at each location. The initial priors of one subject are shown in Fig. 3. The results of the EM algorithm are the posteriors Pt which are p(zi,t = j|xi,t, μj,t, Σj,t) at each location i of each time point t.
Figure 3.
Axial view of the initial atlas probabilities for Subject II combining a healthy atlas and user defined lesions (spheres).
One advantage of our method is that the number of channels Dt at different time point t can vary, which allows us to use different sets of modalities at each time point, a practice which can occur in clinical scanning, and thus provides flexibility for handling clinical diagnosis.
2.2 Construction of a personalized atlas
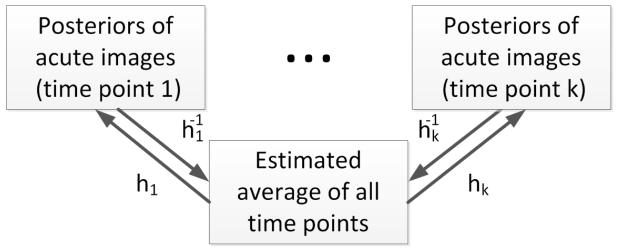
We use the segmentation results (the posteriors) to create personalized atlases using the unbiased diffeomorphic atlas construction method.15 Here we assume that there is no topological changes between the images of different time points. The flowchart of constrction of personalized atlas is depicted in Fig. 4. In atlas construction, we estimate an average (set of probability density functions / PDF) that requires the minimum amount of deformation ht to transform into the posteriors Pt at every time point t, specifically:
| (2) |
where D(e, ht) is the distance of deformations ht to the identity transform e. We use these average PDF as tissue prior probability maps in subsequent segmentations to get more consistent results by combining information from all time points.
Figure 4.
Illustration of construction of personalized atlas.
The number of classes Kt at different time points in longitudinal TBI images typically varies because bleeding or edema can disappear in follow-up scans. Following,16 we address this problem by combining the posteriors of lesions (j ≥ 5) at a specific time point t. The combined lesion class formed by follows our observation that both hemorrhagic and non-hemorrhagic lesions in preceding scans can still be observed in subsequent scans, though they may change appearance due to recovery. Compared to deformable atlas building using the MR image intensities, the benefit of this approach is that intensity calibration is not needed.17 Fig. 5 shows an example of a constructed personalized atlas.
Figure 5.
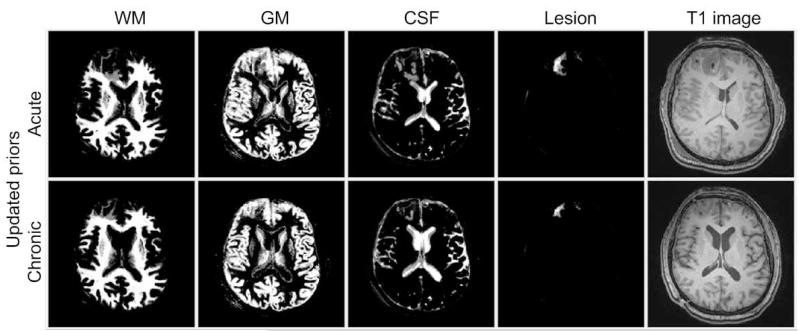
Constructed personalized atlas for Subject II, where the average PDF is deformed to the space at each time point and functions as tissue prior maps.
3. RESULTS
We apply our framework to data sets of three patients with TBI, each with two time points (acute and chronic). The image data of each subject include T1, T2, FLAIR, and GRE modalities. We use manual segmentations by a human expert as ground truth. Also, we compare our result to supervised segmentation (i.e. independent segmentations at each time point). We compare our results and the supervised segmentation results to the ground truth using the Dice coefficient, which is a standard similarity index in the range of 0 to 1 to measure the volumetric overlap of two binary segmentations. Dice coefficient values comparing segmentation results to the ground truth are shown in Table. 1. Please note that the Dice coefficients are relatively low due to the complex boundary shape and relatively random spatial distribution of lesions. The volume of lesions for each subject in the manual segmentation is shown in Table. 2. Our framework generally performs better than independent segmentations, with the primary exception of subject III where there is almost no lesion in the chronic scans and the lesion volumes are very small. The visualization of deformation field via determinant of Jacobian and vector magnitude is shown in Fig. 6. Axial view of acute images of subject I and of the associated segmentation using our framework are presented in Fig. 7.
Table 1.
Dice values comparing automatic segmentation results against ground truth, using temporally independent segmentations and our approach. ANHL is acute non-hemorrhagic lesion, AHL is acute hemorrhagic lesion, CL is chronic lesion.
| Dice values | ||||
|---|---|---|---|---|
|
| ||||
| Lesion types | ANHL | AHL | CL | |
|
| ||||
| I | 0.5080 | 0.4672 | 0.2455 | |
| Indepedent analysis | II | 0.2165 | 0.3550 | 0.4899 |
| III | 0.2747 | 0.2500 | 0.1945 | |
|
| ||||
| I | 0.5990 | 0.5962 | 0.3435 | |
| Joint analysis | II | 0.4398 | 0.5938 | 0.6637 |
| III | 0.4768 | 0.2764 | 0.1176 | |
Table 2.
Lesion volumes (in voxels) for each subject in the manual segmentations. ANHL is acute non-hemorrhagic lesion, AHL is acute hemorrhagic lesion, CL is chronic lesion.
| Subject | ANHL | AHL | CL |
|---|---|---|---|
| I | 57297 | 24486 | 9678 |
| II | 57526 | 36820 | 8847 |
| III | 50151 | 18136 | 1060 |
Figure 6.
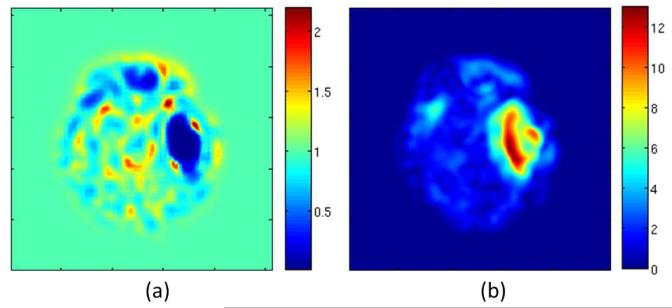
Visualization of the deformation field of Subject I via Jacobian determinant (a) and vector magnitude (b).
Figure 7.
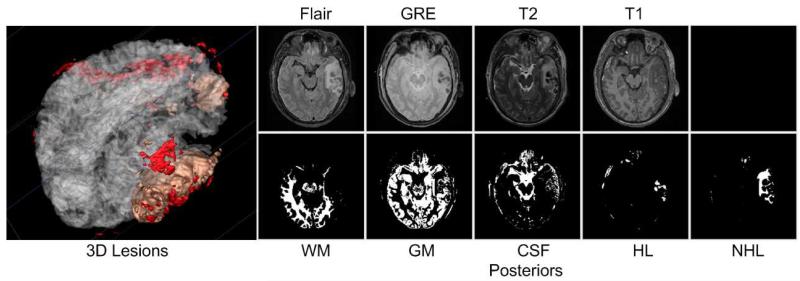
Results of our method for the acute images of Subject I. NHL is non-hemorrhagic lesion, HL is hemorrhagic lesion.
4. CONCLUSIONS
In this paper, we presented a segmentation framework for longitudinal TBI images. The framework is initialized through manual input describing primary lesion sites at each time point, which are then refined by a joint approach composed of Bayesian segmentation and construction of a personalized atlas. Our proposed framework has the advantage of being able to deal with different sets of modalities at each time point. The proposed joint analysis of different time points yields improved results compared to independent analysis.
There are several limitations in our proposed method. One limitation of the proposed method is that we assume there are no topological changes in the images of different time points so that we can use diffeomorphic atlas construction method to build personlized atlas. However, for longitudinal images of TBI patients this assumption is not always true. One possible solution is to mask the lesion area before registration.18 Alternatively, registration methods which are robust to missing correspondence could be used.19-22 Moreover, the approach will have limited applicability when lesion volumes are very small. In this case, the proposed joint approach may not be able to segment and capture these structures.
In the future, we would like to model the topological changes and thus make the atlas construction robust to topological changes. We will explore potential application of our method to aid registration of longitudinal MR images, with TBI, such as the geometric metamorphosis method proposed by Niethammer et al.23 We intend to apply our method to quantify recovery from longitudinal brain MR images with TBI under different treatments, with the potential of determining effective treatment strategies in the future.
ACKNOWLEDGMENTS
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149.
REFERENCES
- [1].Faul M, Xu L, Wald MM, Coronado VG. Traumatic Brain Injury in the United States: Emergency Department Visits, Hospitalizations and Deaths 2002-2006. Centers for Disease Control and Prevention, National Center for Injury Prevention and Control; Atlanta, GA: 2010. [Google Scholar]
- [2].Irimia A, Chambers MC, Alger JR, Filippou M, Prastawa M, Wang B, Hovda DA, Gerig G, Toga AW, Kikinis R, Vespa PM, Horn JDV. Comparison of acute and chronic traumatic brain injury using semi-automatic multimodal segmentation of MR volumes. J. Neurotrauma. 2011 Nov;28 doi: 10.1089/neu.2011.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Thatcher RW, Camacho M, Salazar A, Linden C, Biver C, Clarke L. Quantitative mri of the gray-white matter distribution in traumatic brain injury. J. Neurotrauma. 1997 Jan;14 doi: 10.1089/neu.1997.14.1. [DOI] [PubMed] [Google Scholar]
- [4].Leemput KV, Maes F, Vandermeulen D, Suetens P. Automated model-based tissue classification of mr images of the brain. IEEE Trans. Med. Imaging. 1999:897–908. doi: 10.1109/42.811270. [DOI] [PubMed] [Google Scholar]
- [5].Zhang Y, Brady M, Smith SM. Segmentation of brain mr images through a hidden markov random field model and the expectation maximization algorithm. IEEE Trans. Med. Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- [6].Tu Z, Narr K, Dollár P, Dinov ID, Thompson PM, Toga AW. Brain anatomical structure segmentation by hybrid discriminative/generative models. IEEE Trans. Med. Imaging. 2008;27(4):495–508. doi: 10.1109/TMI.2007.908121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Prastawa M, Bullitt E, Ho S, Gerig G. A brain tumor segmentation framework based on outlier detection. Medical Image Analysis. 2004;8(3):275–283. doi: 10.1016/j.media.2004.06.007. [DOI] [PubMed] [Google Scholar]
- [8].Prastawa M, Bullit E, Moon N, Leemput K, Gerig G. Automatic brain tumor segmentation by subject specific modification of atlas priors. Academic Radiology. 2003;10:1341–1348. doi: 10.1016/s1076-6332(03)00506-3. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Clark MC, Hall LO, Goldgof DB, Velthuizen R, Reed F, Silbiger MS. Automatic tumor segmentation using knowledge-based techniques. IEEE Trans. Med. Imaging. 1998;17:187–201. doi: 10.1109/42.700731. [DOI] [PubMed] [Google Scholar]
- [10].Ho S, Bullitt E, Gerig G. Level-set evolution with region competition: Automatic 3-d segmentation of brain tumors. ICPR (1) 2002:532. [Google Scholar]
- [11].Menze BH, Leemput KV, Lashkari D, Weber M-A, Ayache N, Golland P. A generative model for brain tumor segmentation in multi-modal images. MICCAI (2) 2010:151–159. doi: 10.1007/978-3-642-15745-5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the em algorithm. JOURNAL OF THE ROYAL STATISTICAL SOCIETY, SERIES B. 1977;39(1):1–38. [Google Scholar]
- [13].Mclachlan GJ, Krishnan T. The EM Algorithm and Extensions. Wiley-Interscience; 1997. [Google Scholar]
- [14].Lloyd SP. Least squares quantization in pcm. IEEE Transactions on Information Theory. 1982;28:129–137. [Google Scholar]
- [15].Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. NeuroImage. 2004;23(Supplement 1):S151–S160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- [16].Ha L, Prastawa M, Gerig G, Gilmore J, Silva C, Joshi S. Efficient probabilistic and geometric anatomical mapping using particle mesh approximation on GPUs. Int J Biomed Imaging. 2011;2011 doi: 10.1155/2011/572187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lorenzen P, Prastawa M, Davis B, Gerig G, Bullitt E, Joshi SC. Multi-modal image set registration and atlas formation. Medical Image Analysis. 2006;10:440–451. doi: 10.1016/j.media.2005.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bretta M, Leff AP, Rorden C, Ashburner J. Spatial normalization of brain images with focal lesions using cost function masking. Neuroimage. 2001 Aug;14:486–500. doi: 10.1006/nimg.2001.0845. [DOI] [PubMed] [Google Scholar]
- [19].Periaswamy S, Farid H. Medical image registration with partial data. Medical Image Analysis. 2006:452–464. doi: 10.1016/j.media.2005.03.006. [DOI] [PubMed] [Google Scholar]
- [20].Chitphakdithai N, Duncan JS. Non-rigid registration with missing correspondences in preoperative and postresection brain images. MICCAI (1)’10. 2010:367–374. doi: 10.1007/978-3-642-15705-9_45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ou Y, Sotiras A, Paragios N, Davatzikos C. Dramms: Deformable registration via attribute matching and mutual-saliency weighting. Medical Image Analysis. 2011;15(4):622–639. doi: 10.1016/j.media.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Li X, Long X, Wyatt C. Registration of images with topological change via Riemannian embedding. ISBI. 2011:1247–1252. [Google Scholar]
- [23].Niethammer M, Hart GL, Pace DF, Vespa PM, Irimia A, Horn JDV, Aylward SR. Geometric metamorphosis. MICCAI (2) 2011:639–646. doi: 10.1007/978-3-642-23629-7_78. [DOI] [PMC free article] [PubMed] [Google Scholar]