Figure 6. ULD of TsUCH37 binding to ubiquitin.
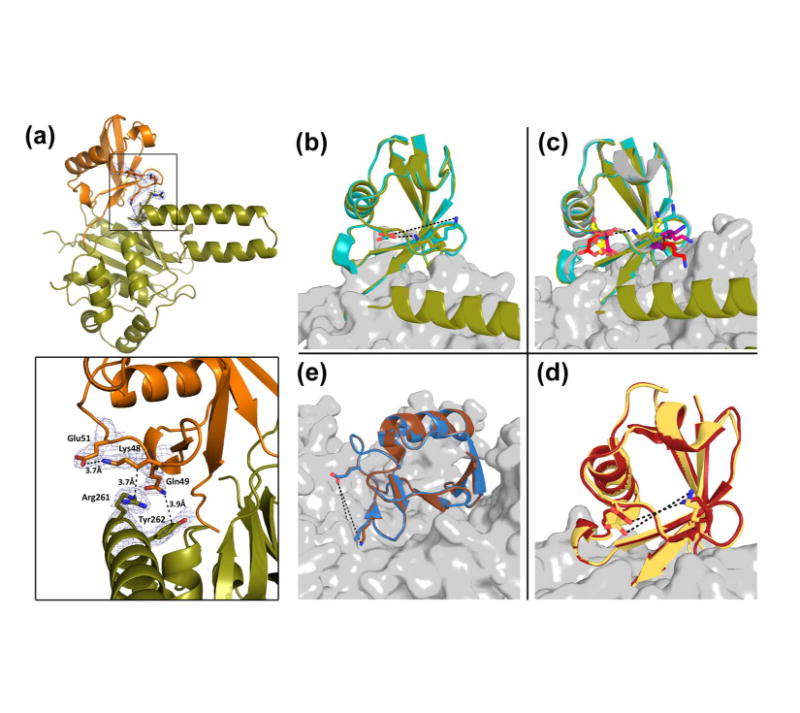
(a) TsUCH37ΔC46 (in olive) ULD residues interacting with UbVME (in orange). Inset shows Arg261 and Tyr262 interactions with UbVME, as well as the intra-molecular salt bridge formed between Lys48 and Glu51 of ubiquitin. Density rendered from 2Fo-Fc map contoured to 0.7σ. (b-e) Comparison of the Lys48-Glu51 distance in ubiquitin observed in other DUB-ubiquitin structures. (b) Lys48 and Glu51 form a 3.7Å salt bridge in TsUCH37ΔC46 – UbVME structure (olive), but not in TsUCH37cat-UbVME structure (9.9 Å). (c) The same distance in all other UCH-ubiquitin structures are ≥9Å: UCHL3-UbVME in yellow (PDB ID 1XD3), UCHL1-UbVME in red (PDB ID 3KW5), Yuh1-Ubal in pink (PDB ID 1CMX), PfUCHL3-UbVME in purple (PDB ID 2WDT), TsUCH37cat-UbVME in teal. (d) The same distance in OTU-ubiquitin structures: Otu1-ubiquitin in dark red (PDB ID 3BY4) is 8.7 Å, and in DUBA-Ubal in pale yellow (PDB ID 3TMP) is 6.0 Å. (e) The same distance in HAUSP/USP7-Ubal (PDB ID 1NBF in blue) is 10Å and in USP14-Ubal (PDB ID 2AYO in brown) is 10.9Å.
