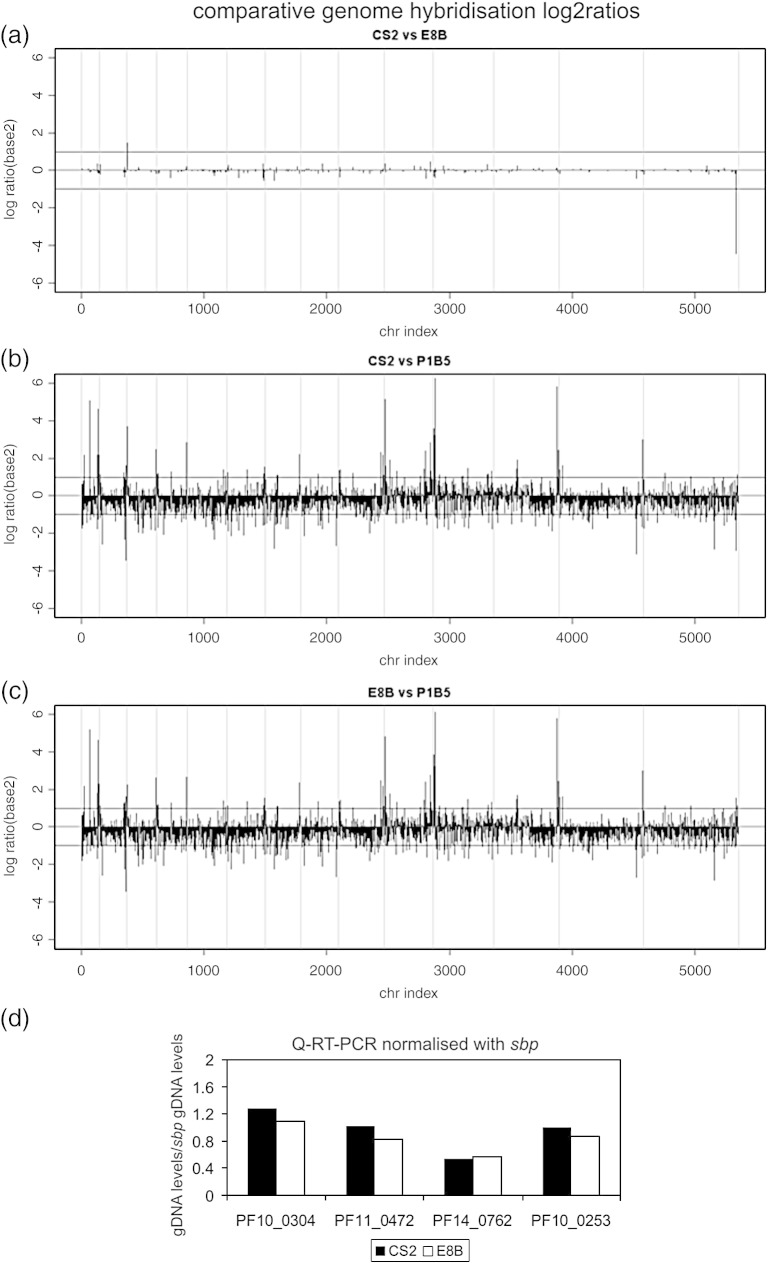
Fig. 3.
Comparative genome hybridisation log2ratios of (a) CS2 compared to E8B, (b) CS2 compared to P1B5, and (c) E8B compared to P1B5. The genes (vertical black bars) are plotted as physical order on the x-axis whilst the log ratios are on the y-axis. The zero line shows the distribution of the genes if no variation was to be found between the two strains compared. The horizontal lines at + 1 and − 1 are limits indicating amplification (log2ratio > 1) and deletion (log2ratio < − 1), respectively. The grey vertical bars delimit the chromosomes. (d) The skeleton binding protein gene sbp and genes identified as possibly deleted or duplicated by microarray were quantitated in both CS2 and E8B gDNA using standard curves of serially diluted gDNA. The levels of sbp were used to normalise the levels of the other genes so that the resulting relative values compare the level of each gene in equivalent amounts of CS2 and E8B gDNA.