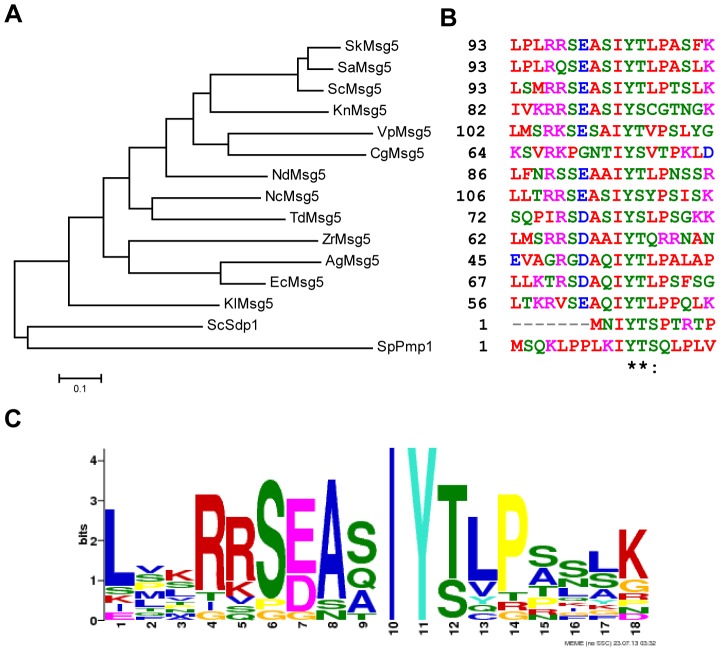
Figure 1. Conservation and consensus pattern of the IYT motif of yeast MKPs.
(A) Dendrogram of S. cerevisiae (Sc) full sequences of genes encoding MKPs Msg5 and Sdp1 and the orthologous sequences from the indicated yeast species. (B) ClustalW multiple sequence alignment of the region containing the IYT motif within MKPs from Saccharomyces kudriavzevii (Sk), Saccharomyces arboricola (Sa), Naumovozyma dairenensis (Nd), Naumovozyma castellii (Nc), Torulaspora delbrueckii (Td), Kazachstania naganishii (Kn), Vanderwaltozyma polyspora (Vp), Zygosaccharomyces rouxii (Zr), Ashbya gossypii (Ag), Kluyveromyces lactis (Kl) Candida glabrata (Cg), Schizosaccharomyces pombe (Sp) and Eremothecium cymbalariae (Ec). Dendrogram was created with the software Mega 5.2. Conserved residues are dotted while identical residues are labelled with asterisks. (C) Consensus pattern and sequence logo of the IYT motif generated by the NEME software [38]. The overall height of each stack indicates the sequence conservation at that position (measured in bits), whereas the height of symbols within the stack reflects the relative frequency of the corresponding amino acid at that position.